Response, 🪙🎲🎰🐜🗡️🪖🛡️#
+ Expand
Analysis
- The Purest Example of Shakespeare’s Poetic Drama
Unlike later histories, which balance action with introspection, Richard II is almost entirely verse—no prose, no comic relief, no distracting subplots. It is Shakespeare at his most elevated, refining blank verse into a lyrical, almost incantatory mode of expression. Richard’s speeches, in particular, are some of the most exquisite poetry in the canon. The play is saturated with metaphor, imagery, and symbolism—so much so that it can feel like a ritualistic meditation on kingship, time, and fate rather than a conventional drama.
Consider Richard’s speech in Act 3, Scene 2:"For God’s sake, let us sit upon the ground
And tell sad stories of the death of kings."- The Most Complex Portrait of Kingship Before Hamlet
Shakespeare builds Richard II around a fundamental political and philosophical question: What makes a king? Richard begins as the divinely ordained ruler, steeped in the medieval belief that kingship is sacred, but by the end of the play, he has been reduced to a mere man. This transition is agonizing and profound, as Shakespeare stages not just a political coup but an existential unraveling.- Psychological and Political Modernity
Richard II dramatizes the performance of power better than any other Shakespearean history. Richard initially appears untouchable, but his rule is exposed as a carefully maintained illusion—his fall from grace is not just a loss of political power but of identity itself. In an age when political legitimacy was shifting from divine right to realpolitik, Shakespeare captures the anxiety of a world in transition.- Richard and Bolingbroke: One of Shakespeare’s Most Fascinating Power Struggles
Unlike the later Henriad plays, where power struggles often play out through military action, Richard II is a battle of words and personas. Bolingbroke represents the practical, Machiavellian future of kingship—he’s adaptable, pragmatic, and understands that power is taken, not given. Richard, by contrast, clings to a fading medieval world of divine rule, seeing himself as a Christlike figure rather than a man who must govern effectively.- The Deposition Scene (Act 4, Scene 1)
This scene alone earns Richard II a place among Shakespeare’s greatest works. Richard’s forced abdication is an extraordinary moment of self-awareness—he plays his own tragedy, turning the deposition into a dramatic performance that both humiliates him and elevates him into something greater than a mere mortal king. His use of mirrors, his obsessive focus on the image of himself as a fallen ruler, and his hypnotic self-destruction are all elements that would later define Shakespeare’s greatest tragic heroes.
Conclusion: A Play of Tragic Majesty
If Richard III is the most theatrical of Shakespeare’s histories, Henry V the most heroic, and Hamlet the most philosophical, Richard II is the most poetic and self-aware. It lacks the battlefield drama of Henry IV and Henry V, but what it offers instead is a devastating meditation on power, identity, and the transformation of political reality. It’s Shakespeare at his most lyrical and his most profound—less a straightforward history than an existential tragedy in disguise.-- Richard II
A Book on Salience, Absurdity, and the Rug of Being
“I am afraid you don’t quite see the moral of the story,” chirped the Linnet, perched on a twig of smug certainty, feathers fluffed with Victorian pomp. “The what?” bellowed the Water-rat, whiskers bristling, splashing in the muck of existence. “The moral.” “You mean the story has a moral?” “Of course,” said the Linnet, preening like a bird stuffed with Dickensian crumbs. “Well, really,” snapped the Water-rat, tail whipping like a metronome to some irreverent tune, “you should’ve warned me. I’d have said ‘Pooh,’ like a proper critic, and spared myself the sermon.” He roared “Pooh!” with gusto, flicked his tail in defiance, and scurried into his burrow of absurdity. The Duck paddled up, all sighs and damp sentiment. “What do you think of the Water-rat?” she asked. “He has his charms, but I weep for any confirmed bachelor.” “I fear I’ve irked him,” admitted the Linnet. “I told him a story with a moral.” “Oh dear,” quacked the Duck, “that’s always a perilous move.” And I quite agree.

Fig. 1 Digital Library. Our color-coded QR code library, with a franchise model for the digital twin, launches next month—exploring struggle, exchange, and consolidation as rhythms of existence.#
Thus closes Oscar Wilde’s The Devoted Friend, a fin-de-siècle jab at Victorian sanctimony wrapped in a syncopated strut—light, irreverent, and mocking moral cathedrals. Wilde, who exited stage left in 1900 alongside Nietzsche, shared a disdain for ponderous systems. Wilde spun fables to skewer earnestness; Nietzsche smashed certainty with aphoristic glee. Neither would care for neuroscience’s tidy brain maps, yet here we weave their spirits into a tale of salience and rhythm. Imagine the salient network—that cingulo-insular maestro—as the Dude’s rug, tying the brain’s chaotic room together: the Pericentral reflex, the Dorsal Frontoparietal gaze, the Lateral Frontoparietal riff, the Medial Frontoparietal mirror, and its own Cingulo-Insular beat. It’s absurd, elegant, and utterly essential.
Ecclesiastes 3:8
A time to love, and a time to hate; a time of war, and a time of peace
Picture the brain as a saloon band, each network a player in a dusty, fin-de-siècle joint. The Pericentral network thumps the bass—boom, a hand yanks from fire, a colony flinches from the lash. It’s the Water-rat’s “Pooh!”—raw, instinctive, no moral attached. Wilde would grin; Nietzsche might call it the will’s pulse. The Dorsal Frontoparietal network scans the room, hawk-eyed, spotting the nonself—hot iron, foreign flag, a thief in crow’s feathers. It’s the Linnet’s prim focus, certain yet oblivious to its own farce. Nietzsche’s overman stares; Wilde smirks at the pose.
The Lateral Frontoparietal network improvises, riffing on gray notes—self versus nonself, tribal roots clashing with imposed codes. It’s the Duck, teary yet droll, wading through ambiguity. Wilde thrives here; Nietzsche sees choice reborn. The Medial Frontoparietal network croons inward—memory, identity, the “I” forged in Africa’s forge or Shakespeare’s quill. It’s the Linnet’s moral clinging to itself, ripe for Wilde’s scoff and Nietzsche’s hammer. Then, the Cingulo-Insular network—the salient rug—ties it all up, syncing reflex to gaze, riff to croon, a bandleader juggling chaos into rhythm. In biology, it filters threats; in history, it’s Africa blending scars and roots. Wilde might call it a dandy’s flourish—absurdly perfect; Nietzsche hears Dionysian harmony, wild and free.
The Dude’s Rug; LLM
Pericentral. Friendship; Grammar
Dorsal. Miller; Witness 👂🏾
Lateral. Devotion; Agent
Medial. Hans, Wheelbarrow, Miller; Verbs
Cingulo-Insular. Time-is-Broke; Object

Fig. 2 Salience: Cingulo-Insular Network. What tied the room together, dude?#
Take Wilde’s trio: the Water-rat’s “Pooh!” is the Pericentral flinch, shunning the moral’s nonself. The Linnet’s prattle is Dorsal fixation, smugly blind. The Duck’s tears sway in Lateral limbo, half-judging, half-laughing. The Medial hums in the Linnet’s selfhood, until the Cingulo-Insular rug pulls it taut: the Duck’s quack—“that’s always a perilous move”—lands the jest. Nietzsche chuckles; Africa mirrors it—Pericentral recoils, Dorsal eyes, Lateral blends, Medial grasps—all woven by a salient hope, yet absurdly unresolved.
No moral lurks here. Wilde and Nietzsche would bolt at the notion. The salient network isn’t a preacher; it’s the Dude’s rug, tying the brain’s mess into a tumbling weeds rhythm that mocks its own weave. Victorian cadence meets Lebowski’s strut—a dance through absurdity, from Wilde’s wit to Nietzsche’s chaos to Africa’s saga. Five networks, five threads, one rug: no answers, just the beat.
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['DNA, RNA, 5%', 'Peptidoglycans, Lipoteichoics', 'Lipopolysaccharide', 'N-Formylmethionine', "Glucans, Chitin", 'Specific Antigens'],
'Voir': ['PRR & ILCs, 20%'],
'Choisis': ['CD8+, 50%', 'CD4+'],
'Deviens': ['TNF-α, IL-6, IFN-γ', 'PD-1 & CTLA-4', 'Tregs, IL-10, TGF-β, 20%'],
"M'èléve": ['Complement System', 'Platelet System', 'Granulocyte System', 'Innate Lymphoid Cells, 5%', 'Adaptive Lymphoid Cells']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['PRR & ILCs, 20%'],
'paleturquoise': ['Specific Antigens', 'CD4+', 'Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'],
'lightgreen': ["Glucans, Chitin", 'PD-1 & CTLA-4', 'Platelet System', 'Innate Lymphoid Cells, 5%', 'Granulocyte System'],
'lightsalmon': ['Lipopolysaccharide', 'N-Formylmethionine', 'CD8+, 50%', 'TNF-α, IL-6, IFN-γ', 'Complement System'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights
def define_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
('PRR & ILCs, 20%', 'CD8+, 50%'): '20/80',
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD8+, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
('CD8+, 50%', 'PD-1 & CTLA-4'): '80/20',
('CD8+, 50%', 'Tregs, IL-10, TGF-β, 20%'): '95/5',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD4+', 'PD-1 & CTLA-4'): '20/80',
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Platelet System'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Granulocyte System'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Innate Lymphoid Cells, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Adaptive Lymphoid Cells'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Platelet System'): '1/8',
('PD-1 & CTLA-4', 'Granulocyte System'): '1/7',
('PD-1 & CTLA-4', 'Innate Lymphoid Cells, 5%'): '1/6',
('PD-1 & CTLA-4', 'Adaptive Lymphoid Cells'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Platelet System'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Granulocyte System'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Define edges to be highlighted in black
def define_black_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
}
# Calculate node positions
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
black_edges = define_black_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
edge_colors = []
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
edge_colors.append('black' if (source, target) in black_edges else 'lightgrey')
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color=edge_colors,
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™: Pericentral", fontsize=18)
plt.show()
# Run the visualization
visualize_nn()
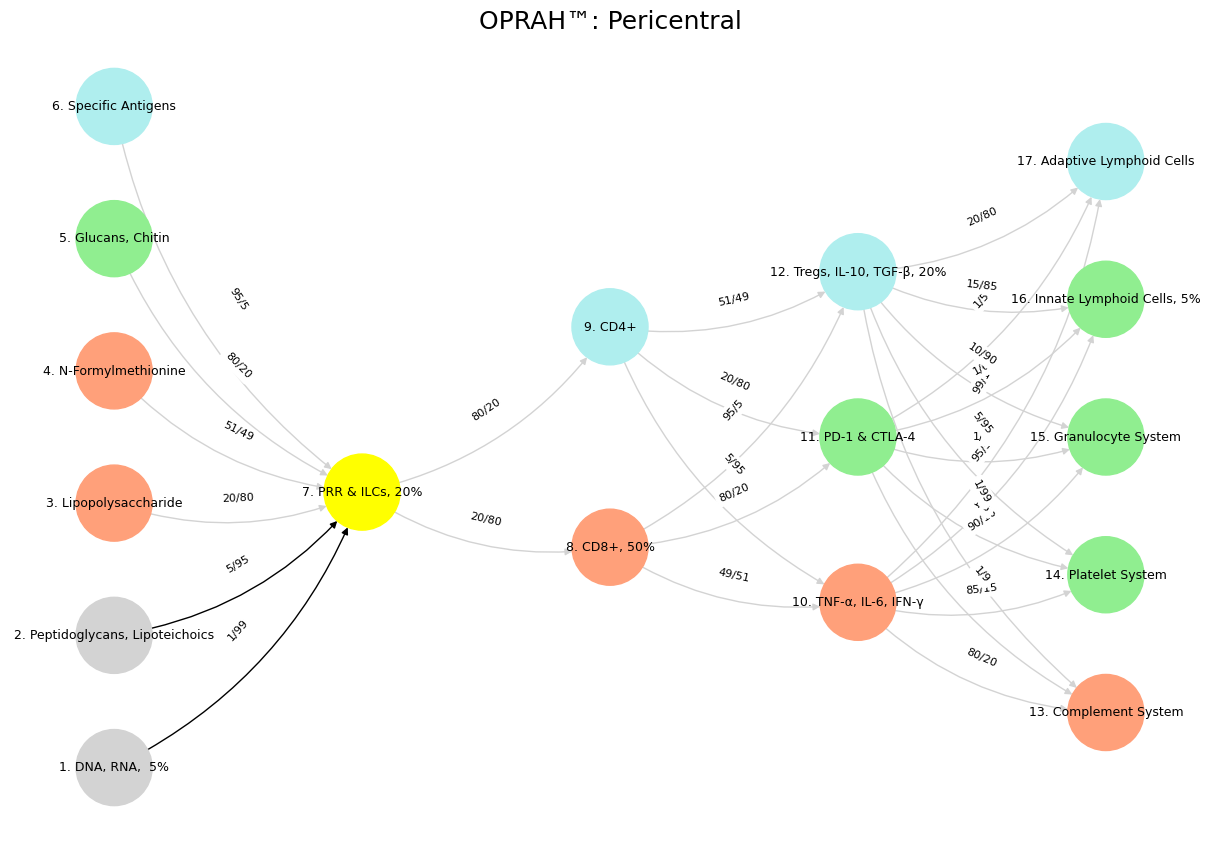

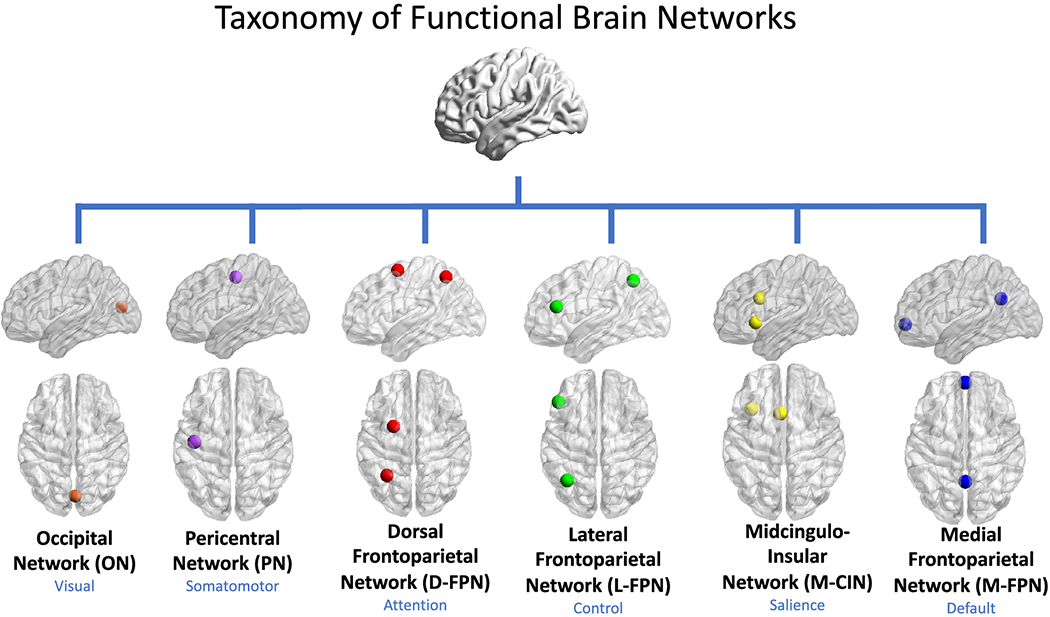
Fig. 3 Taxonomy of functional brain networks. In our proposed taxonomy, networks are referred to by anatomical names that best describe six ubiquitous large-scale functional systems. The names in blue refer to the broad cognitive domains with which a given anatomical system is most commonly associated. Only 1-2 core nodes of each network are depicted here, though it is understood that multiple additional cortical, subcortical, and cerebellar nodes may be affiliated with a given network. Source: Uddin et al#