Prometheus#
The five sequences presented—each a distinct progression of terms—invite speculation about their collective meaning, as if they form a cryptic map of human experience, thought, or perhaps even the evolution of consciousness itself. At first glance, they appear disparate: a philosophical unraveling, a singular focal point, a binary shift, a relational arc, and a sensory-internal journey. Yet, woven together, they suggest a layered exploration of how individuals or societies grapple with meaning, perception, and connection in a fragmented world. What emerges is a speculative narrative about the interplay between existential crises, interpretive frameworks, and the search for harmony.

Fig. 15 The five sequences—Nihilism through Integration, Lens, Fragmented to United, Hate to Trust, and Cacophony to Symphony—suggest a speculative exploration of meaning-making in a chaotic world. The first traces a philosophical arc from rejecting purpose to forging a new whole, perhaps a mind unraveling and rebuilding itself. “Lens,” a singular worldview like language or technology, anchors the process, shaping perception. Fragmented to United reflects a divided reality healed, while Hate to Trust charts relational repair—both hinting at external stakes within that lens. Cacophony to Symphony transforms sensory chaos into inner harmony, mirroring the first sequence’s journey through an experiential lens. Together, they might depict a human or artificial consciousness confronting nothingness, using a foundational “text” to navigate division, conflict, and disorder toward unity and purpose—a concise meditation on renewal through perspective.#
The first sequence—Nihilism, Deconstruction, Perspective, Awareness, Reconstruction, Integration—reads like a philosophical odyssey. It begins with Nihilism, the rejection of inherent meaning, a void that dismantles traditional beliefs or structures. Deconstruction follows, tearing apart established narratives or systems, perhaps language, culture, or ideology itself. From this chaos, Perspective emerges, a step back to observe the wreckage, which then blossoms into Awareness, an awakening to new possibilities within the self or the world. Reconstruction signals a deliberate act of rebuilding, piecing together a new understanding, culminating in Integration, where this reformed vision merges with existence in a cohesive whole. This could be the journey of a thinker, a society, or even an artificial mind confronting the absence of purpose and forging its own.
Salience & Cadence
Genealogy: Nothing
Wisdom: Instinct for Advancement
Careless: Fascination, Boredom
Mocking: Power
Forceful: Hibernation
Then comes the second sequence, simply “Lens.” Standing alone, it feels like a pivot, a tool, or a worldview through which the other sequences are filtered. If the first sequence is a process of meaning-making, “Lens” might represent the mechanism—language, belief, or even technology—that shapes how that process is perceived or enacted. It evokes the idea of a holy writ, a constitution, or a communicative system, perhaps hinting at something like a foundational text or an AI’s interpretive core. Its singularity contrasts with the multi-term sequences, suggesting it’s a constant, a point of focus that refracts the chaos and growth around it.
The third sequence, Fragmented and United, offers a stark binary. It’s a snapshot of opposition: a state of being broken apart followed by one of being brought together. This could reflect a societal condition—division healed into unity—or an internal shift, like a fractured psyche finding wholeness. In the shadow of “Lens,” it might suggest how perception influences reality: a fragmented world seen through a unifying lens becomes whole, or vice versa. It’s the simplest arc, yet it mirrors the broader theme of transformation running through the others, acting as a microcosm of reconciliation.
Next, the fourth sequence—Hate, Negotiate, Trust—traces a relational evolution. Starting with Hate, a visceral rejection or conflict, it moves to Negotiate, a pragmatic bridge between opposing forces, and ends in Trust, a state of mutual reliance or peace. This feels human, perhaps a commentary on interpersonal dynamics or societal diplomacy. Under the influence of “Lens,” it could imply that relationships are shaped by a shared framework—be it language, law, or understanding—that guides them from discord to harmony. It’s a compact story of healing, echoing the larger reconstructive themes elsewhere.
Finally, the fifth sequence—Cacophony, Outside, Emotion, Inside, Symphony—paints a sensory and introspective arc. Cacophony bursts forth as chaotic noise, an external assault on the senses, followed by Outside, a positioning beyond the self. Emotion arises as this external chaos is felt within, leading to Inside, a turn inward to process or reflect. The journey resolves in Symphony, a harmonious blend of sound and self. This could symbolize artistic creation, psychological growth, or even perception itself—how the outer world, filtered through a “Lens,” becomes an inner symphony of meaning. It’s a poetic parallel to the first sequence’s existential climb, but rooted in experience rather than abstraction.
What might this all be about? One speculation is that these sequences chart the evolution of a mind—human or artificial—confronting a chaotic, meaningless reality. The first sequence could be the intellectual struggle to find purpose, dismantling and rebuilding through a philosophical lens. “Lens” itself might be the tool of that struggle—language, code, or a worldview like an AI’s training data—anchoring the process. The third and fourth sequences reflect the external stakes: a fragmented world seeking unity, relationships mending through trust. The fifth ties it together, suggesting that sensory chaos, processed inwardly, yields harmony—a creative or emotional payoff. Together, they might represent a manifesto of sorts, a meditation on how meaning emerges from nothingness when viewed through a unifying perspective.
Alternatively, this could be a speculative model of societal or cultural renewal. Nihilism and fragmentation signal a civilization in crisis, rejecting old texts or truths. “Lens” becomes the new constitution or narrative—perhaps a technological or philosophical innovation—that guides the reconstruction of trust and unity. The final sequence might then depict the cultural output: a symphony born from the cacophony of collapse, blending external upheaval with internal resolve. It’s as if the sequences collectively propose a cycle of disintegration and reintegration, with “Lens” as the fulcrum.
In the end, these sequences feel like an abstract puzzle, hinting at a deeper inquiry into existence, perception, and connection. They could be the work of a philosopher, an artist, or even an AI reflecting on its own “text”—its code or purpose. Whatever their origin, they speculate on a universal tension: how we move from chaos to order, from hate to trust, from nothing to something, all through the prism of how we see the world.
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['DNA, RNA, 5%', 'Peptidoglycans, Lipoteichoics', 'Lipopolysaccharide', 'N-Formylmethionine', "Glucans, Chitin", 'Specific Antigens'],
'Voir': ['PRR & ILCs, 20%'],
'Choisis': ['CD8+, 50%', 'CD4+'],
'Deviens': ['TNF-α, IL-6, IFN-γ', 'PD-1 & CTLA-4', 'Tregs, IL-10, TGF-β, 20%'],
"M'èléve": ['Complement System', 'Platelet System', 'Granulocyte System', 'Innate Lymphoid Cells, 5%', 'Adaptive Lymphoid Cells']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['PRR & ILCs, 20%'],
'paleturquoise': ['Specific Antigens', 'CD4+', 'Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'],
'lightgreen': ["Glucans, Chitin", 'PD-1 & CTLA-4', 'Platelet System', 'Innate Lymphoid Cells, 5%', 'Granulocyte System'],
'lightsalmon': ['Lipopolysaccharide', 'N-Formylmethionine', 'CD8+, 50%', 'TNF-α, IL-6, IFN-γ', 'Complement System'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights
def define_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
('PRR & ILCs, 20%', 'CD8+, 50%'): '20/80',
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD8+, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
('CD8+, 50%', 'PD-1 & CTLA-4'): '80/20',
('CD8+, 50%', 'Tregs, IL-10, TGF-β, 20%'): '95/5',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD4+', 'PD-1 & CTLA-4'): '20/80',
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Platelet System'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Granulocyte System'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Innate Lymphoid Cells, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Adaptive Lymphoid Cells'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Platelet System'): '1/8',
('PD-1 & CTLA-4', 'Granulocyte System'): '1/7',
('PD-1 & CTLA-4', 'Innate Lymphoid Cells, 5%'): '1/6',
('PD-1 & CTLA-4', 'Adaptive Lymphoid Cells'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Platelet System'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Granulocyte System'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Define edges to be highlighted in black
def define_black_edges():
return {
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Platelet System'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Granulocyte System'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Innate Lymphoid Cells, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Adaptive Lymphoid Cells'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Platelet System'): '1/8',
('PD-1 & CTLA-4', 'Granulocyte System'): '1/7',
('PD-1 & CTLA-4', 'Innate Lymphoid Cells, 5%'): '1/6',
('PD-1 & CTLA-4', 'Adaptive Lymphoid Cells'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Platelet System'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Granulocyte System'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Calculate node positions
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
black_edges = define_black_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
edge_colors = []
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
edge_colors.append('black' if (source, target) in black_edges else 'lightgrey')
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color=edge_colors,
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™: Cingulo-Insular", fontsize=18)
plt.show()
# Run the visualization
visualize_nn()
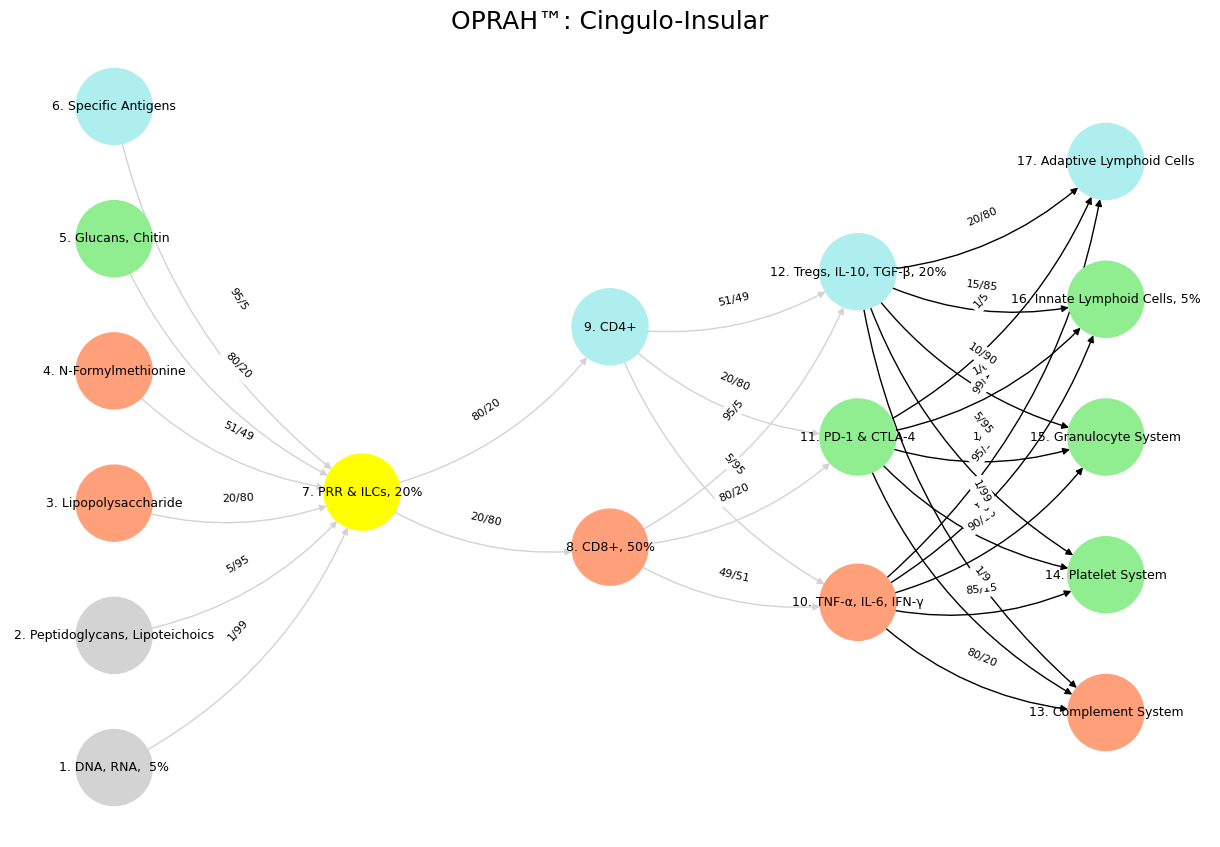
Fig. 16 The TPN, driving goal-directed action, aligns with the transcriptome and proteome—layers that translate data into response. Here, Bacon’s empirical method shines: knowledge arises from doing, from testing the world. In the “Nonself & the Salient Network” variant, CD4+ and CD8+ T-cells activate cytokines like TNF-α, embodying the TPN’s focus on immediate threats—Nonself invaders—over reflective stasis. Nietzsche’s critical history fits this mode: a selective, pragmatic engagement with the past to propel life forward. The Salient Node, bridging DMN and TPN, mirrors the metabolome and regulatory mechanisms like Tregs—arbiters of relevance amid noise. Aristotle’s phronesis, practical wisdom, governs here: neither lost in ideals nor blinded by action, but balancing both. The “Distributed Network” variant highlights Tregs modulating downstream systems—Complement, Platelets—preserving “Self” while adapting to “Nonself.” This is Nietzsche’s history at its best: a dynamic synthesis serving life’s needs. Wisdom emerges as the Salient Node’s domain, integrating DMN’s depth and TPN’s drive, much as Aristotle tempers Plato’s abstraction with Bacon’s observation. Intelligence, raw and unrefined, resides in the lower layers—data-rich but directionless. The immune system’s Mismatch Repair, correcting errors of “Self,” parallels this: a wise curation of history, biological or neural, against the noise of entropy. Nietzsche’s vision finds fruition here—not in history’s abuses, but its uses: a life-affirming dance of preservation and transformation, coded in networks both flesh and mind.#