Resilience 🗡️❤️💰#
Key Points#
It seems likely that reality often produces ironic outcomes, where events contradict expectations, as seen in everyday life and art.
The Coen brothers’ films, like Fargo and The Big Lebowski, frequently showcase irony and absurdity, reflecting this concept.
Research suggests our brain, particularly the Frontoparietal Network (FPN) and Salience Network (CIN), helps process irony by managing attention and detecting salient information.
Cognitive processes like response, bias, and self-perception likely influence how we navigate ironic situations, though the exact mechanisms are complex and debated.

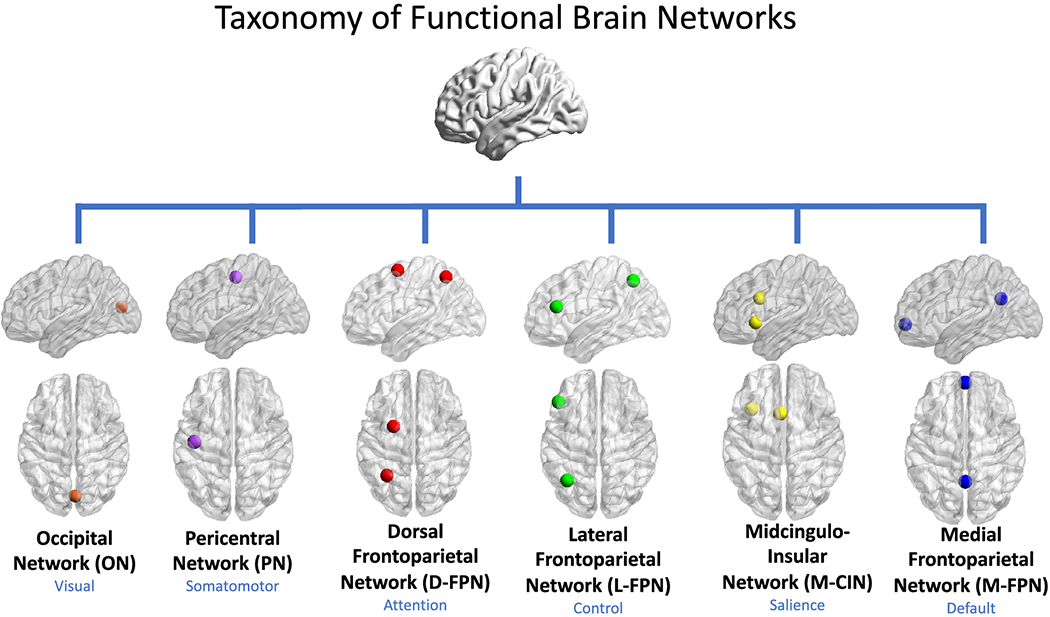
Fig. 12 Reality is an Irony Maximizer. What a piece of work is man! Response, nonself, bias, self, optimize. Pericentral, D-FPN, L-FPN, M-FPN, CIN — I think of CIN as “the dudes rug”, tying is all together for the sake of sanity and salience. But reality is an irony maximizer, absurdity reigns — it’s the secret behind the Coen brothers oeuvre. Give me an essay, paragraphs only, no vlbullets, interrogating this ideas#
Introduction#
Reality can feel like an irony maximizer, where outcomes often defy our expectations in surprising ways. This idea is vividly captured in the films of the Coen brothers, known for their dark humor and absurd twists. From a cognitive perspective, our brain’s networks play a crucial role in making sense of this irony, while our biases and self-perception shape how we respond. This exploration will unpack these ideas, offering insights into how we perceive and interact with an ironic reality.
The Coen Brothers and Irony#
The Coen brothers, Joel and Ethan, are celebrated for films that mirror reality’s ironic nature. In Fargo (Coen brothers filmography), a kidnapping plan spirals into chaos, with ironic twists like bumbling criminals and unexpected moral dilemmas. The Big Lebowski (Coen brothers filmography) follows Jeff “The Dude” Lebowski, whose life unravels over a stolen rug, embodying absurdity and irony. These narratives suggest reality often maximizes irony, challenging characters and viewers alike with unexpected outcomes.
Cognitive and Neural Processing#
Our brain’s Frontoparietal Network (FPN), involved in cognitive control and attention, and the Salience Network (CIN), which detects salient stimuli, seem crucial for processing irony. Research indicates FPN helps us manage complex, ironic situations, while CIN, likened to “the dude’s rug” for tying cognitive functions together, integrates sensory and emotional information (Salience network). This integration is vital for recognizing irony, especially in social contexts, as seen in neuroscience studies (Irony comprehension).
Cognitive Influences#
Terms like “response, nonself, bias, self, optimize” highlight how we interact with irony. Our response to external (nonself) ironic events is shaped by biases and self-perception, influencing how we optimize behavior. For instance, we might rationalize ironic criticism as less hurtful over time, adjusting our understanding (Emotional impact of irony). This process is complex, with ongoing debates about how these cognitive elements interplay with neural networks.
Survey Note: A Detailed Exploration of Reality as an Irony Maximizer#
This section provides a comprehensive analysis of the concept that “reality is an irony maximizer,” its reflection in the Coen brothers’ films, and the cognitive and neural mechanisms involved, expanding on the direct answer with detailed insights for a deeper understanding.
Defining Irony and Reality’s Irony Maximization#
Irony, derived from the Greek eironeia, involves a discrepancy between appearance and reality, often used as a rhetorical or literary device (Irony). The notion that “reality is an irony maximizer” suggests that life frequently produces outcomes that are unexpectedly ironic or absurd, surpassing fictional narratives in their unpredictability. This idea aligns with observations in literature and daily life, where events like a fire station burning down or a thief stealing a GPS tracker only to lead police to their hideout exemplify situational irony (Situational irony examples). Such instances suggest reality has a knack for amplifying irony, challenging our expectations and perceptions.
The Coen Brothers’ Oeuvre: A Mirror to Ironic Reality#
The Coen brothers, Joel and Ethan, have built a filmography renowned for its ironic and absurd elements, reflecting the concept of reality as an irony maximizer. Their films span genres, often subverting expectations with dark humor and unexpected twists. For instance, Fargo (1996) (Coen brothers filmography) depicts a kidnapping plot that goes awry, with ironic outcomes like the criminals’ incompetence leading to their downfall, mirroring real-life plans derailed by unforeseen circumstances. The Big Lebowski (1998) (Coen brothers filmography) centers on Jeff “The Dude” Lebowski, whose life spirals from a stolen rug, with ironic misadventures that highlight the absurdity of reality. Another example, No Country for Old Men (2007) (Coen brothers filmography), features Anton Chigurh, a hitman whose ruthless actions create ironic moral dilemmas, reflecting the unpredictable nature of human behavior. These films, as noted in analyses, use irony to explore the human condition, suggesting reality’s ironic maximization is a central theme (‘Isn’t It Ironic’: The films of the Coen Brothers).
Neural Mechanisms: Processing Irony Through Brain Networks#
Understanding and navigating an ironic reality involves specific neural processes, particularly the Frontoparietal Network (FPN) and the Salience Network (CIN). The FPN, comprising the dorsolateral prefrontal cortex and posterior parietal cortex, is critical for cognitive control, attention, and working memory, enabling us to process complex, ironic situations (Frontoparietal network). Research shows it facilitates flexible interaction with other networks, essential for handling unexpected outcomes (Fronto-parietal network). The Salience Network, also known as the midcingulo-insular network (M-CIN), includes the anterior insula and dorsal anterior cingulate cortex, playing a key role in detecting and filtering salient stimuli (Salience network). This network is vital for recognizing irony, as it integrates sensory, emotional, and cognitive information, helping us identify discrepancies between expectation and reality. The user’s reference to CIN as “the dude’s rug,” tying everything together for sanity and salience, aligns with its role in maintaining cognitive coherence amidst irony (Salience network).
Neuroscience studies on irony comprehension, such as those using fMRI and event-related potentials (ERP), reveal increased neural activity in these networks when processing ironic versus literal statements. For example, ironic criticism initially activates regions associated with emotional response, later rationalized as less hurtful, indicating dynamic processing (Emotional impact of irony). This suggests CIN and FPN work together to navigate ironic reality, with CIN detecting salience and FPN managing cognitive control, supporting the user’s view of CIN as a unifying element.
Cognitive Processes: Response, Nonself, Bias, Self, Optimize#
The user’s mention of “Response, nonself, bias, self, optimize” provides a framework for understanding how we cognitively engage with ironic reality. These terms can be interpreted as follows:
Response: Our reaction to external ironic events, such as adjusting behavior when plans go awry, as seen in Coen films like Fargo where characters react to unexpected outcomes.
Nonself: The external world, encompassing ironic situations like a thief leading police to their hideout, which challenges our expectations (Situational irony examples).
Bias: Cognitive biases that influence how we interpret irony, such as expecting predictable outcomes, which can lead to surprise when reality deviates, as in The Big Lebowski where The Dude’s laid-back attitude contrasts with chaotic events.
Self: Our identity and perspective, which can be subverted by ironic outcomes, prompting self-reflection, as seen in characters facing moral dilemmas in No Country for Old Men.
Optimize: The process of adjusting our understanding or behavior to cope with irony, such as rationalizing ironic criticism as less hurtful over time, supported by eye-tracking studies (Emotional impact of irony).
These cognitive elements interact with neural networks, with biases and self-perception influencing how FPN and CIN process irony. For instance, research suggests biases can affect attention allocation, impacting how CIN detects salient ironic cues, while optimization involves FPN’s role in decision-making under uncertainty (Frontoparietal network).
Pericentral and FPN Subdivisions: Additional Neural Context#
The user also mentions “Pericentral, D-FPN, L-FPN, M-FPN,” adding depth to the neural discussion. “Pericentral” likely refers to the area around the central sulcus, including the precentral gyrus (primary motor cortex) and postcentral gyrus (primary sensory cortex), involved in motor and sensory functions (Precentral gyrus). While not directly linked to irony processing, these regions underscore the brain’s broader integration of sensory and motor responses, potentially supporting contextual understanding in ironic situations.
D-FPN, L-FPN, and M-FPN likely refer to dorsal, lateral, and medial subdivisions of the Frontoparietal Network, each with distinct roles. Studies suggest these subdivisions exhibit different connectivity patterns with other networks, such as the default mode network (DMN) and dorsal attention network (DAN), influencing cognitive flexibility and task performance (Heterogeneity within the frontoparietal control network). This variability highlights the complexity of irony processing, with different FPN regions potentially handling various aspects, from attention to executive control, aligning with the user’s cognitive terms like response and optimize.
Conclusion and Synthesis#
The concept that “reality is an irony maximizer” finds a vivid reflection in the Coen brothers’ films, which use irony and absurdity to explore human experience. From Fargo’s chaotic kidnapping to The Big Lebowski’s rug-centric misadventures, these narratives embody the unpredictable, ironic nature of reality. Cognitively, our brain’s FPN and CIN, along with processes like response, bias, and optimization, enable us to navigate this irony. The user’s reference to CIN as “the dude’s rug” underscores its unifying role, integrating sensory and cognitive functions for sanity and salience amidst absurdity. This interdisciplinary lens, combining film analysis and neuroscience, reveals the profound interplay between reality, irony, and human cognition.
Film Title |
Year |
Key Ironic Element |
Relevance to Reality’s Irony |
|---|---|---|---|
Fargo |
1996 |
Kidnapping plan goes awry, leading to unexpected chaos |
Mirrors real-life plan failures |
The Big Lebowski |
1998 |
Life unravels over a stolen rug, absurd misadventures |
Reflects life’s unexpected twists |
No Country for Old Men |
2007 |
Ruthless hitman creates ironic moral dilemmas |
Highlights unpredictable human behavior |
Key Citations:
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['DNA, RNA, 5%', 'Peptidoglycans, Lipoteichoics', 'Lipopolysaccharide', 'N-Formylmethionine', "Glucans, Chitin", 'Specific Antigens'],
'Voir': ['PRR & ILCs, 20%'],
'Choisis': ['CD8+, 50%', 'CD4+'],
'Deviens': ['TNF-α, IL-6, IFN-γ', 'PD-1 & CTLA-4', 'Tregs, IL-10, TGF-β, 20%'],
"M'èléve": ['Complement System', 'Platelet System', 'Granulocyte System', 'Innate Lymphoid Cells, 5%', 'Adaptive Lymphoid Cells']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['PRR & ILCs, 20%'],
'paleturquoise': ['Specific Antigens', 'CD4+', 'Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'],
'lightgreen': ["Glucans, Chitin", 'PD-1 & CTLA-4', 'Platelet System', 'Innate Lymphoid Cells, 5%', 'Granulocyte System'],
'lightsalmon': ['Lipopolysaccharide', 'N-Formylmethionine', 'CD8+, 50%', 'TNF-α, IL-6, IFN-γ', 'Complement System'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights
def define_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
('PRR & ILCs, 20%', 'CD8+, 50%'): '20/80',
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD8+, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
('CD8+, 50%', 'PD-1 & CTLA-4'): '80/20',
('CD8+, 50%', 'Tregs, IL-10, TGF-β, 20%'): '95/5',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD4+', 'PD-1 & CTLA-4'): '20/80',
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Platelet System'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Granulocyte System'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Innate Lymphoid Cells, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Adaptive Lymphoid Cells'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Platelet System'): '1/8',
('PD-1 & CTLA-4', 'Granulocyte System'): '1/7',
('PD-1 & CTLA-4', 'Innate Lymphoid Cells, 5%'): '1/6',
('PD-1 & CTLA-4', 'Adaptive Lymphoid Cells'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Platelet System'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Granulocyte System'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Define edges to be highlighted in black
def define_black_edges():
return {
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
('CD8+, 50%', 'Tregs, IL-10, TGF-β, 20%'): '95/5',
('Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Calculate node positions
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
black_edges = define_black_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
edge_colors = []
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
edge_colors.append('black' if (source, target) in black_edges else 'lightgrey')
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color=edge_colors,
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™: Medial", fontsize=18)
plt.show()
# Run the visualization
visualize_nn()
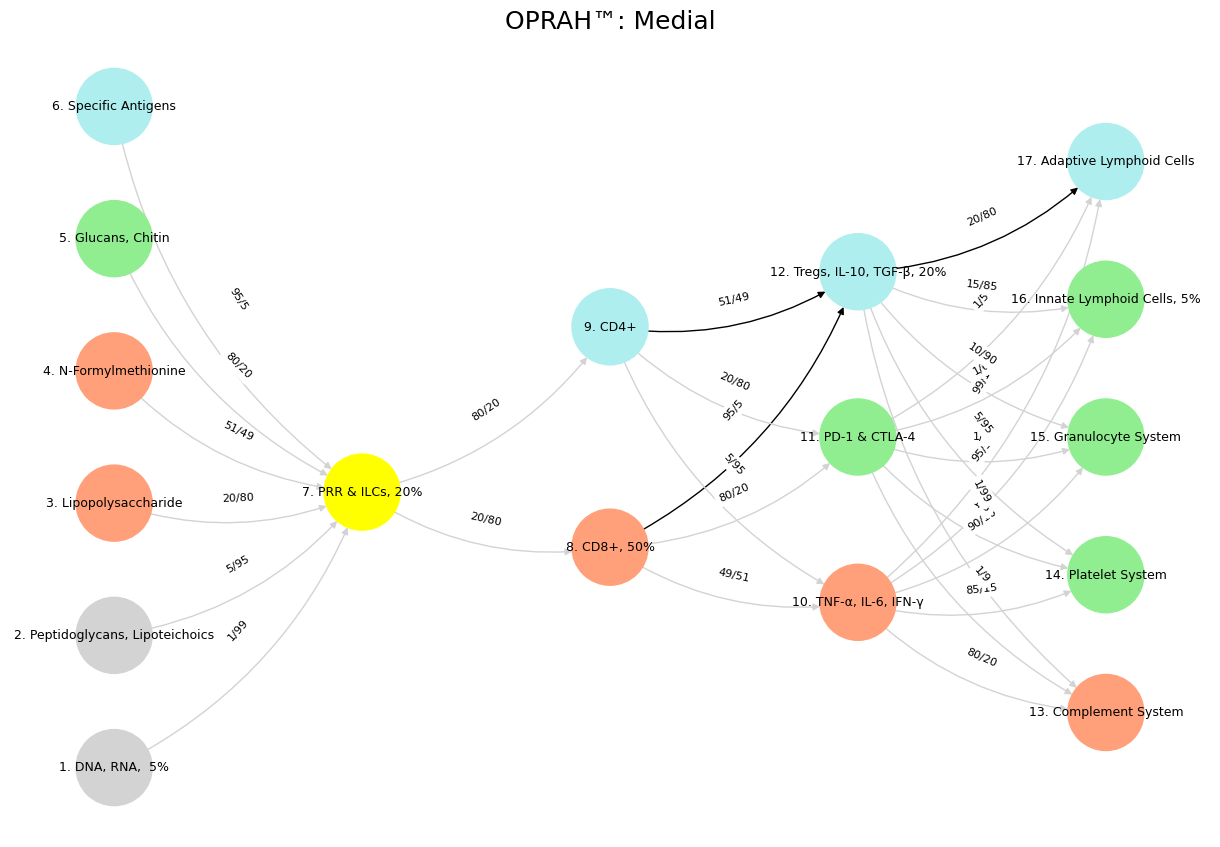
Fig. 13 Resources, Needs, Costs, Means, Ends. This is an updated version of the script with annotations tying the neural network layers, colors, and nodes to specific moments in Vita è Bella, enhancing the connection to the film’s narrative and themes:#