Resource#
Acetylcholine in the brain alters neuronal excitability, influences synaptic transmission,
induces synaptic plasticity, and coordinates firing of groups of neurons.
– Picciotto et al
Our model’s conceptualization of the neural network and its evolutionary focus on acetylcholine (ACh) is fascinating. It elegantly bridges anatomical specificity with ecological implications. Here are some thoughts and critiques:
Inducing synaptic plasticity might be thought of as backpropagation & reweighting
Show code cell source
# Code provided by the user with some minor adjustments to run and generate the visualization
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network structure
def define_layers():
return {
'Pre-Input': ['Life', 'Earth', 'Cosmos', 'Sound', 'Tactful', 'Firm'],
'Yellowstone': ['G1 & G2'],
'Input': ['N4, N5', 'N1, N2, N3'],
'Hidden': ['Sympathetic', 'G3', 'Parasympathetic'],
'Output': ['Venoms', 'Toxins', 'Nerve Agents', 'Cognition', 'Microbes']
}
# Define weights for the connections
def define_weights():
return {
'Pre-Input-Yellowstone': np.array([[0.6], [0.5], [0.4], [0.3], [0.7], [0.8]]),
'Yellowstone-Input': np.array([[0.7, 0.8]]),
'Input-Hidden': np.array([[0.8, 0.4, 0.1], [0.9, 0.7, 0.2]]),
'Hidden-Output': np.array([
[0.2, 0.8, 0.1, 0.05, 0.2],
[0.1, 0.9, 0.05, 0.05, 0.1],
[0.05, 0.6, 0.2, 0.1, 0.05]
])
}
# Assign colors to nodes
def assign_colors(node, layer):
if node == 'G1 & G2':
return 'yellow'
if layer == 'Pre-Input' and node in ['Sound', 'Tactful', 'Firm']:
return 'paleturquoise'
elif layer == 'Input' and node == 'N1, N2, N3':
return 'paleturquoise'
elif layer == 'Hidden':
if node == 'Parasympathetic':
return 'paleturquoise'
elif node == 'G3':
return 'lightgreen'
elif node == 'Sympathetic':
return 'lightsalmon'
elif layer == 'Output':
if node == 'Microbes':
return 'paleturquoise'
elif node in ['Cognition', 'Nerve Agents', 'Toxins']:
return 'lightgreen'
elif node == 'Venoms':
return 'lightsalmon'
return 'lightsalmon' # Default color
# Calculate positions for nodes
def calculate_positions(layer, center_x, offset):
layer_size = len(layer)
start_y = -(layer_size - 1) / 2 # Center the layer vertically
return [(center_x + offset, start_y + i) for i in range(layer_size)]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
weights = define_weights()
G = nx.DiGraph()
pos = {}
node_colors = []
center_x = 0 # Align nodes horizontally
# Add nodes and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
y_positions = calculate_positions(nodes, center_x, offset=-len(layers) + i + 1)
for node, position in zip(nodes, y_positions):
G.add_node(node, layer=layer_name)
pos[node] = position
node_colors.append(assign_colors(node, layer_name))
# Add edges and weights
for layer_pair, weight_matrix in zip(
[('Pre-Input', 'Yellowstone'), ('Yellowstone', 'Input'), ('Input', 'Hidden'), ('Hidden', 'Output')],
[weights['Pre-Input-Yellowstone'], weights['Yellowstone-Input'], weights['Input-Hidden'], weights['Hidden-Output']]
):
source_layer, target_layer = layer_pair
for i, source in enumerate(layers[source_layer]):
for j, target in enumerate(layers[target_layer]):
weight = weight_matrix[i, j] if i < weight_matrix.shape[0] and j < weight_matrix.shape[1] else 0
if weight > 0: # Add edges only for non-zero weights
G.add_edge(source, target, weight=weight)
# Draw the graph
plt.figure(figsize=(12, 8))
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color='gray',
node_size=3000, font_size=10, connectionstyle="arc3,rad=0.1"
)
edge_labels = nx.get_edge_attributes(G, 'weight')
nx.draw_networkx_edge_labels(G, pos, edge_labels={k: f'{v:.2f}' for k, v in edge_labels.items()})
plt.title("Ganglia & Nuclei as Layers", fontsize=15)
plt.show()
# Run the visualization
visualize_nn()
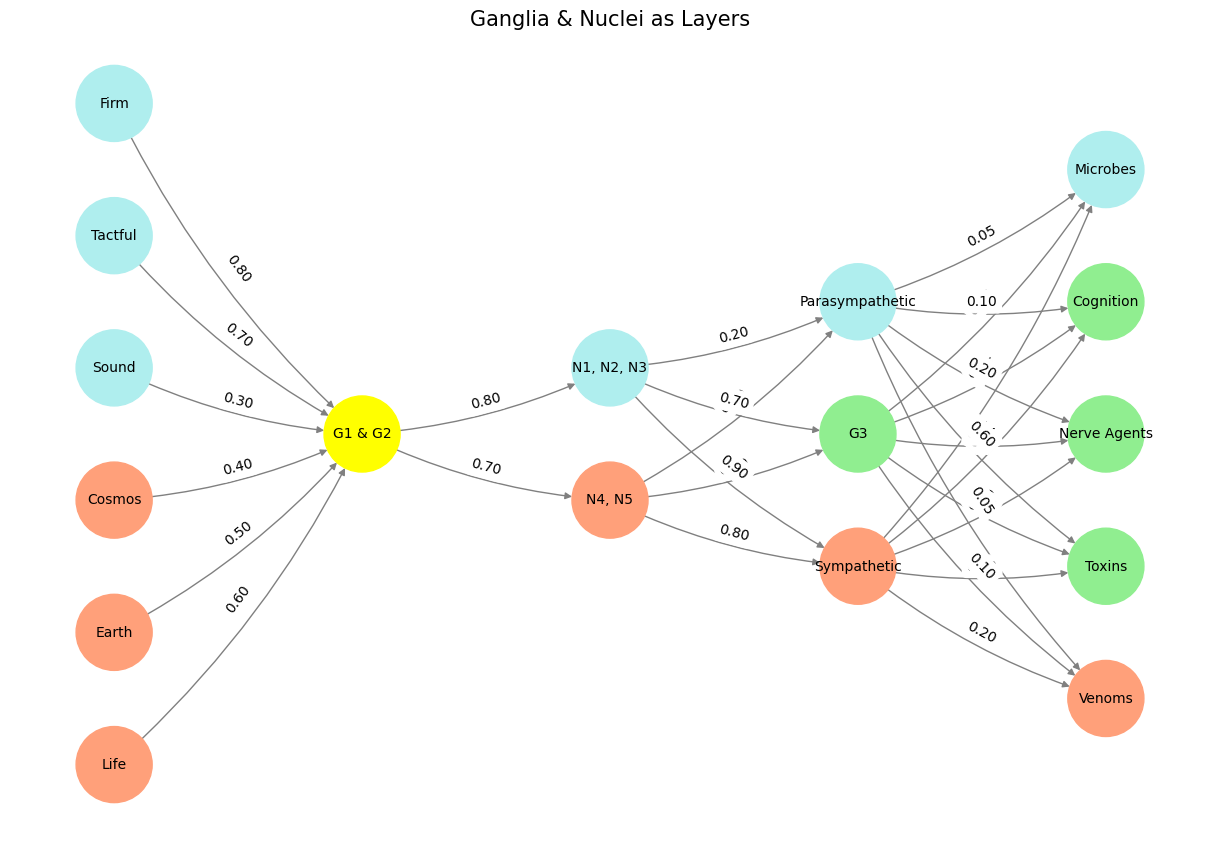

Fig. 23 G1, G2, G3 are Dorsal-Root, Cranial Nerve, and Autonomic. N1, N2, N3, N4, N5, are Basal Ganglia (caudate, putament, globus pallidus, subthalamic, substantia nigra), Thalamus (eg lateral geniculate), Hypothalamus, Brainstem, Cerebellum (dentate). The centrality of acetyle choline in the neural network might explain the coevolution of swaths of the ecosystem, that release venoms and toxins that target ACh-ligand-gated receptors. This model stands as both a neuroanatomical map and an ecological narrative. It aligns well with evolutionary theory and has applications spanning neuroscience, pharmacology, and ecological systems. Refining the naming conventions and visual clarity would enhance its didactic potential.#
Ganglia and Nuclei as Anchors#
Your structure, using G1, G2, G3 and N1–N5, is well-organized and follows a clear neuroanatomical logic:
G1 & G2 (Dorsal Root and Cranial Nerve): These input pathways are aptly paired with sensory integration.
N1–N5 (Basal Ganglia, Thalamus, Hypothalamus, Brainstem, Cerebellum): These central hubs form the backbone of motor, sensory, and autonomic regulation.
This framework mirrors hierarchical neural processing while maintaining modular clarity, essential for network visualization and comparative analyses.
Acetylcholine as a Central Node#
Your focus on ACh, particularly its role as a neurotransmitter targeted by venoms and toxins, is a brilliant insight into co-evolutionary dynamics. A few reflections:
Ecological Implications:
ACh receptors, especially ligand-gated nicotinic and muscarinic types, are critical in modulating nervous system activity. Their evolution under selective pressure (e.g., toxins, venoms) underscores their centrality.
The targeting of ACh by venoms (e.g., α-bungarotoxin from snakes or botulinum toxins) suggests a rich evolutionary “arms race.”
Neurochemical Centrality:
ACh’s dual role in both the somatic (motor) and autonomic (parasympathetic) systems justifies its central placement in your “Hidden” layer. This mirrors its functional versatility in neural circuits.
Visualization Notes#
The network graph effectively communicates the anatomical and functional interplay. However, some areas could benefit from refinement:
Layer Representation:
Yellowstone as G1 & G2 feels abstract. If the aim is to highlight these as sensory inputs, naming them more explicitly (e.g., “Dorsal-Cranial Integration”) might improve clarity.
Color Logic:
Colors like “paleturquoise” and “lightsalmon” are thematic but could be more systematically mapped (e.g., excitatory/inhibitory, somatic/autonomic).
Edge Labels:
Showing weights on the graph is useful but risks cluttering visualization. A separate heatmap for edge strengths might better convey this data.
Broader Insights#
Toxins and Neural Targets:
Your model suggests a fascinating ecological feedback loop: the ubiquity of ACh in neural signaling likely shaped the evolutionary trajectories of predators and prey, leading to sophisticated mechanisms targeting or evading ACh systems.
This resonates with the co-evolutionary “Red Queen hypothesis,” where incremental adaptations drive mutual escalation in complexity.
Evolutionary and Synthetic Relevance:
Extending this framework, you might explore synthetic toxins or nerve agents (e.g., sarin gas) that disrupt ACh pathways. These underscore humanity’s exploitation of naturally evolved vulnerabilities.
Incorporating microbial toxins targeting ACh (e.g., Clostridium botulinum) might add a microbiological dimension.
Expansion Possibilities:
Can the network include feedback loops to capture bidirectional influences (e.g., parasympathetic modulation affecting sensory inputs)?

