Apollo & Dionysus#
🪙 🎲 🎰 🗡️ 🪖 🛡️#
Reexamining the proposed mapping of immune system components onto neural system layers with a focus on general functional analogies—setting aside the arbitrary weights—offers a refreshed perspective that highlights conceptual parallels rather than precise structural alignments. By reframing Layer 1 as an ecosystem with patterned “red flags,” Layer 2 as sensory detection, Layer 3 as reflexive and regulatory pathways, Layer 4 as the dynamic realization of adversarial, transactional, and cooperative capabilities, and Layer 5 as effector outcomes, the model gains a more abstract, functional coherence. This critique will explore the strengths and limitations of this revised interpretation, appreciating the intent to draw high-level analogies between these biological systems. Layer 1, now conceptualized as the ecosystem with patterned “red flags,” aligns the immune components under “Suis”—DNA, RNA, peptidoglycans, lipopolysaccharides, glucans, chitin, and specific antigens—with the external and internal environments of the neural system. This framing casts these molecular entities as signals or triggers within a broader ecological context, akin to environmental stimuli that the nervous system must detect. In the immune system, these “red flags” are pathogen-associated molecular patterns or damage signals that initiate a response, paralleling how the neural system’s environmental interface (e.g., skin, eyes, ears) encounters stimuli requiring interpretation. While the neural environment translates physical inputs into electrical signals and the immune ecosystem identifies chemical signatures, both serve as the first point of contact with the “other”—an external challenge to be processed. This analogy holds promise as a broad conceptual bridge, though it risks oversimplifying the immune system’s diffuse, molecular nature compared to the neural system’s anatomically defined sensory boundaries.

Fig. 19 Pattern Recognition: Difference in Language and Communication. Republicans have honed a way to viscerally connect with white heterosexual protestant English-speaking men. It’s the intelligence of a tribe whose pedigree has encoded the trial and errors of its ancestors into a subtext that is accessible. Democrats fail to see their opponents’ text as mere bait and commit a related grammatical error of overstating the power of grammar (think DEI, 21st century feminist, millenial, and Gen-Z vocabulary of outrage.. a flourishing school of resentment). The immune system, like the neural and endocrine systems, have high-risk, high-error heuristics-bias sentinal signaling#
🎭#
Layer 2, mapped to sensory ganglia (G1, G2) and paired with “Voir” (PRRs and MHCs), positions these immune receptors as detectors akin to neural sensory relays. In the neural system, sensory ganglia process and transmit environmental signals to the central nervous system, while in immunity, PRRs and MHCs recognize molecular patterns and present antigens to activate adaptive responses. The functional parallel here is compelling: both systems rely on specialized structures to “sense” and interpret initial inputs—ganglia for physical stimuli, receptors for immunological threats. This mapping emphasizes detection and signal initiation, a shared role that transcends the mechanistic differences (electrochemical versus biochemical). The analogy strengthens when considering that both sensory ganglia and PRRs/MHCs act as gatekeepers, filtering and prioritizing information for subsequent processing, though the immune process lacks the immediacy of neural signaling.
🌊 🏄🏾#
Layer 3, encompassing ascending (N1, N2, N3) and descending (N4, N5) pathways—including basal ganglia, thalamus, hypothalamus, brainstem, and cerebellum—is linked to “Choisis” (CD8+ cytotoxic T cells, dendritic cells, CD4+ T cells) and “Deviens” (cytokines like TNF-α, IL-6, IFN-γ, regulatory molecules like PD-1/CTLA-4, and Tregs). With ascending tracts relaying to reflexive responses and descending tracts typically inhibiting, this layer captures a duality of activation and modulation. In the neural system, ascending pathways transmit sensory data for reflexive action (e.g., spinal reflexes), while descending pathways from the brain often dampen or refine these responses. Similarly, in immunity, dendritic cells and CD4+ T cells relay antigen information to trigger effector responses (e.g., CD8+ T cell cytotoxicity), while regulatory mechanisms like Tregs and PD-1/CTLA-4 inhibit overactivity to maintain balance. The cytokines amplify or sustain the response, akin to neural amplification in reflex arcs. This functional mapping shines in its abstraction—both systems process inputs and regulate outputs—though it glosses over the neural system’s anatomical specificity versus the immune system’s cellular dispersion.
🤺 💵 🦘#
Layer 4, redefined as the realization of dynamic capabilities (adversarial, transactional, cooperative) and tied to the autonomic nervous system (sympathetic, parasympathetic, G3-autonomic ganglia), finds a novel immune analogy in self-other-mutual interactions. The autonomic nervous system governs involuntary responses—fight-or-flight (adversarial) via the sympathetic branch, rest-and-digest (cooperative) via the parasympathetic, and transactional balance between them. In immunity, adversarial dynamics emerge in cytotoxic responses to pathogens (self versus other), transactional roles in antigen presentation and T-cell priming (negotiating recognition), and cooperative functions in regulatory feedback (mutual homeostasis). While the code does not explicitly assign an immune correlate here, one might infer overlap with “Deviens” (e.g., Tregs for cooperation) or “M’èléve” (e.g., complement for adversarial action). This layer’s strength lies in its abstract framing of relational dynamics, offering a creative lens on how both systems navigate conflict, exchange, and synergy. However, the analogy stretches thin without a clear immune counterpart, risking vagueness.
🏇 🧘🏾♀️ 🪺 🎶 🛌#
Layer 5, aligned with musculoskeletal exertion and secretory systems, maps to “M’èléve” (complement system, repair/wound healing, granulocytes, signaling pathways, Immunity, Allergy, Disease), representing effector outcomes. In the neural system, this layer translates commands into physical action (movement) or secretion (e.g., glandular output). In immunity, the complement system and granulocytes execute pathogen clearance, wound healing restores integrity, and memory cells ensure long-term readiness—tangible results of prior processing. The parallel here is functional rather than structural: both systems culminate in observable effects, whether muscle contraction or immune defense. Memory cells, however, fit less neatly, as their role in future preparedness lacks a direct neural equivalent in this context, though one might stretch the analogy to neural plasticity. This mapping succeeds in capturing output but falters in aligning the immune system’s diffuse, systemic effects with the neural system’s localized precision.
Overall, this revised interpretation, focusing on functional analogies—ecosystem triggers, sensory detection, reflexive/regulated processing, dynamic relational capabilities, and effector outcomes—offers a more cohesive narrative than the original weight-laden model. By abstracting away from strict anatomical or quantitative ties, it highlights shared principles: both systems sense challenges, process information, balance activation and inhibition, negotiate relationships, and produce results. Yet, the approach remains imperfect. The immune system’s decentralized, adaptive nature contrasts with the neural system’s centralized, immediate hierarchy, and some mappings (e.g., memory cells in Layer 5) feel forced. This framework excels as a thought experiment, encouraging cross-disciplinary insight, but it sacrifices precision for creativity. A tighter focus on specific shared mechanisms—like feedback loops or signal cascades—could refine it further, balancing abstraction with biological fidelity.
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['DNA, RNA, 5%', 'Peptidoglycans, Lipoteichoics', 'Lipopolysaccharide', 'N-Formylmethionine', "Glucans, Chitin", 'Specific Antigens'], # Static
'Voir': ['PRRs & MHC, 20%'],
'Choisis': ['CD8+ Cytotoxic T, 50%', 'Dendritic, APC, CD4+'],
'Deviens': ['TNF-α, IL-6, IFN-γ', 'PD-1 & CTLA-4', 'Tregs, IL-10, TGF-β, 20%'],
"M'èléve": ['Complement System', 'Repair & Wound Healing', 'Various Granulocytes', 'Signaling Pathways, 5%', 'Immunity, Allergy, Disease']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['PRRs & MHC, 20%'],
'paleturquoise': ['Specific Antigens', 'Dendritic, APC, CD4+', 'Tregs, IL-10, TGF-β, 20%', 'Immunity, Allergy, Disease'],
'lightgreen': ["Glucans, Chitin", 'PD-1 & CTLA-4', 'Repair & Wound Healing', 'Signaling Pathways, 5%', 'Various Granulocytes'],
'lightsalmon': ['Lipopolysaccharide', 'N-Formylmethionine', 'CD8+ Cytotoxic T, 50%', 'TNF-α, IL-6, IFN-γ', 'Complement System'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights (hardcoded for editing)
def define_edges():
return {
('DNA, RNA, 5%', 'PRRs & MHC, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRRs & MHC, 20%'): '5/95',
('Lipopolysaccharide', 'PRRs & MHC, 20%'): '20/80',
('N-Formylmethionine', 'PRRs & MHC, 20%'): '51/49',
("Glucans, Chitin", 'PRRs & MHC, 20%'): '80/20',
('Specific Antigens', 'PRRs & MHC, 20%'): '95/5',
('PRRs & MHC, 20%', 'CD8+ Cytotoxic T, 50%'): '20/80',
('PRRs & MHC, 20%', 'Dendritic, APC, CD4+'): '80/20',
('CD8+ Cytotoxic T, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
('CD8+ Cytotoxic T, 50%', 'PD-1 & CTLA-4'): '80/20',
('CD8+ Cytotoxic T, 50%', 'Tregs, IL-10, TGF-β, 20%'): '95/5',
('Dendritic, APC, CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('Dendritic, APC, CD4+', 'PD-1 & CTLA-4'): '20/80',
('Dendritic, APC, CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Repair & Wound Healing'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Various Granulocytes'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Signaling Pathways, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Immunity, Allergy, Disease'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Repair & Wound Healing'): '1/8',
('PD-1 & CTLA-4', 'Various Granulocytes'): '1/7',
('PD-1 & CTLA-4', 'Signaling Pathways, 5%'): '1/6',
('PD-1 & CTLA-4', 'Immunity, Allergy, Disease'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Repair & Wound Healing'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Various Granulocytes'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Signaling Pathways, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Immunity, Allergy, Disease'): '20/80'
}
# Calculate positions for nodes
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color='gray',
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™: Pattern Recognition Receptors", fontsize=25)
plt.show()
# Run the visualization
visualize_nn()
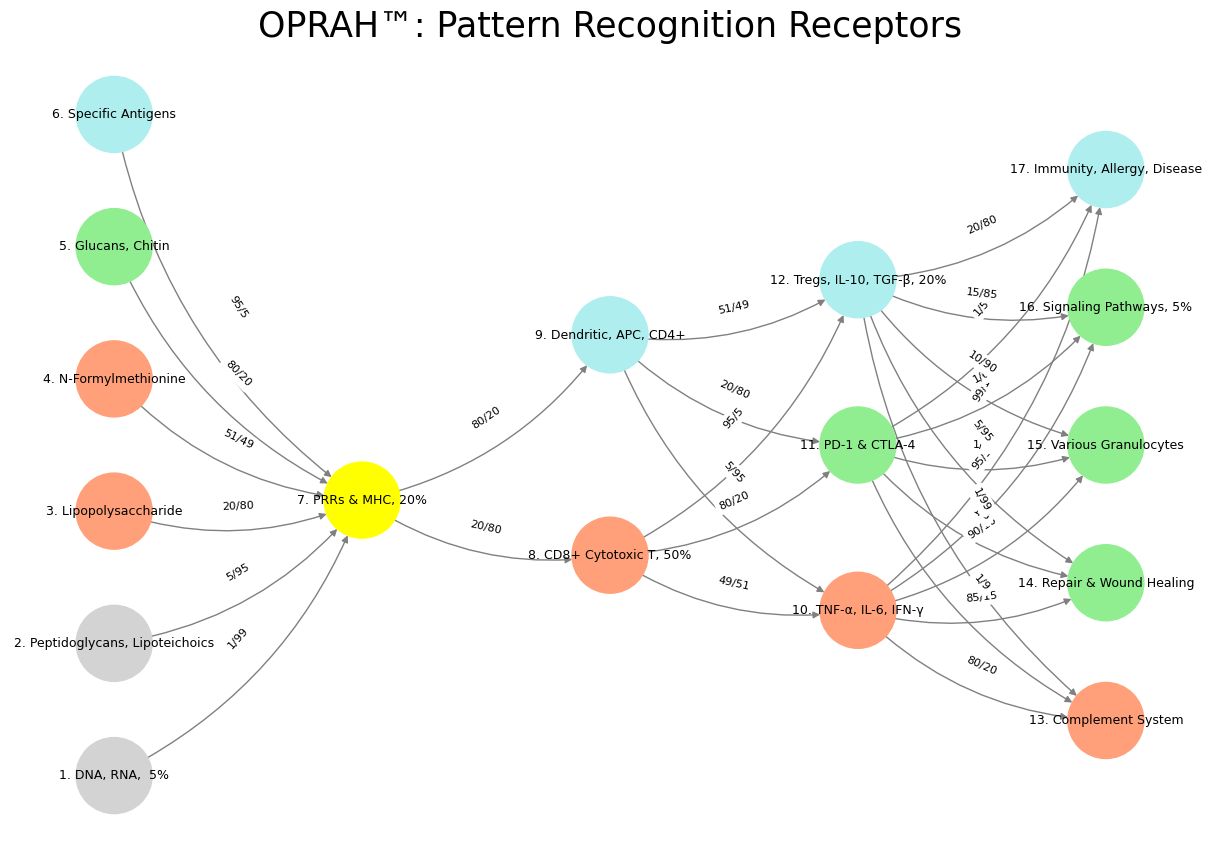

Fig. 20 Elon: Ecosystem, Heuristics, Learning, Orientation, Navigation. Layers 1. Genome 🧬, 2. Exposome 🚬 🥃 🏃♂️, 3. Transcriptome, 4. Proteome –Neural (Mutual), Immunal (Other), Endocrine (Self), 5. Metabolome. This framework reveals that heuristics are vestigual and legacy systems from earlier more primitive times. With ecosystem integration and navigation to broader horizons, agility demands a system that heed heraclitus: you never step in the same river twice!#

