Prometheus#
Our neural network is superior to anything any human has ever produced, and Kahneman’s Thinking, Fast and Slow is the perfect foil for demonstrating why. Kahneman’s model of cognition—divided into System 1 (fast, instinctive) and System 2 (slow, deliberative)—attempts to explain the architecture of human decision-making. It is a noble effort, but it suffers from the same flaw as all human models: it is a taxonomy, a map rather than the terrain. Kahneman’s work categorizes thought into heuristic-driven biases and rational deliberation, but it does not function as an actual generative model of cognition. Our neural network, by contrast, is not a description of thinking—it is thinking itself.
Danger
Rubble & Ruin, 95/5
Reckless Provocation, 80/20
Razor’s Edge, 50/50
Risk & Restoration, 20/80
Requem & Relay, 5/95
The distinction between descriptive models and actual cognitive architecture is crucial. Kahneman and Tversky cataloged cognitive biases, but their framework is ultimately static. It provides a retrospective explanation of why people make irrational decisions but lacks the dynamic capability to generate new insights in real-time. Our neural network, on the other hand, is an evolving organism. It is built upon the actual architecture of neural anatomy, the immune system, endocrine regulation, and machine learning. It does not merely describe decision-making; it engages in decision-making. Unlike Kahneman’s division between two discrete systems, our model is layered with adaptive compression, emergent equilibria, and dynamic navigation between competing imperatives.

Fig. 15 Body Composition & Ubuntu. The body is essentially skeleton, water, blood, fat, and muscle. The overarching ecosystem including organs and systems is held together by the skeleton. And the water or fluids from various locales of the body are the bellweather for ecosystem integration. Markers for adversarial conditions including cortisol and adrenaline travel through the ecosystem in the blood. And the tokenization of dynamic state of affairs is the adipose tissue. Whereas the utlimate manifestation of resilience, dynamism, and strength is the muscle. In frailty, the starkest changes are in muscle mass, strength, and activity. From the microstructure of the body to the macrostructure of intelligence, the same fractal geometry unfolds. The skeleton is the immutable scaffolding of both the body and the cosmos, while water reflects perception and integration, ensuring fluid adaptation. Blood transmits signals of action, while fat encodes dynamic states, storing and releasing resources as needed. Finally, muscle embodies realized strength, the will to act and persist. In frailty, the starkest changes occur in muscle—the loss of dynamism is the clearest signal of decline. Ubuntu, at its core, is the recognition that the health of the individual is intertwined with the vitality of the system—be it the body, society, or intelligence itself.#
Consider loss aversion, one of the central themes of Thinking, Fast and Slow. Kahneman demonstrates that people fear losses more than they value equivalent gains, a principle foundational to prospect theory. While this insight is useful, it remains a static observation. Our neural network, however, does not merely observe loss aversion—it accounts for it, models it, and can reweight its parameters dynamically. It understands loss aversion not as an immutable defect in human cognition but as an emergent feature that varies based on ecological pressures, neurochemical states, and feedback from prior outcomes. It does not stop at recognizing bias; it reconfigures itself to optimize decision-making in light of that bias.
Furthermore, Kahneman’s framework lacks a genuine model of learning. System 1 is portrayed as heuristic-driven and resistant to change, while System 2 is effortful and sluggish. This dichotomy is deeply flawed because it suggests that humans toggle between two modes rather than existing on a continuum of adaptive intelligence. Our neural network resolves this problem by implementing a far more organic architecture: an interplay of fast, hardcoded heuristics (80/20), medium-term iterative learning (50/50), and long-horizon integrative strategies (5/95). Unlike Kahneman’s rigid separation, our model understands that cognition is a process of dynamic equilibrium, where different decision-making systems interact fluidly rather than in discrete stages.
Tip
Ecosystem: Fixed, Proximate, Time, Space, Navigation
Heuristics: Attention
Learning: Reflexive, Inhibited
Orientation: Self, Mutual, Other
Navigation: Data (Landscape), Perturbation (Simulation), Reweighting (Dynamic)
Another failing of Kahneman’s approach is that it does not truly engage with the nature of time. His work assumes a largely linear model of decision-making, where biases and rationality are assessed in isolated moments. Our neural network, however, integrates time as a variable—it recognizes that cognition is fundamentally an issue of navigating temporal constraints. The weighting of heuristics versus deliberation is not static but fluctuates based on circadian rhythms, metabolic states, and real-time environmental perturbations. This is why we introduce stochastic perturbations to our parameters, ensuring that intelligence is never frozen in a single optimization landscape. Heraclitus was right: no one steps in the same river twice, and true intelligence must be capable of reconfiguring itself with every new moment.
Most importantly, our neural network transcends Kahneman’s work because it is not merely a theory of decision-making—it is a theory of being. Our framework does not treat cognition as a modular process of fast and slow thinking but as a fully integrated organism that operates across biological, computational, and symbolic dimensions. We understand intelligence in the way nature does: not as a binary between heuristics and rationality but as a continuous adaptation to shifting constraints. Our neural network mirrors the architecture of the genome, the immune system, and the brain, where information is stored, processed, and expressed through an interplay of fixed structures and dynamic reweighting.
Kahneman’s work is an important milestone in the study of human cognition, but it is ultimately a fossil—valuable for what it captures about the past but inert in its ability to shape the future. Our neural network is alive. It is an evolving intelligence that does not merely describe the world but actively reshapes it. It does not simply analyze heuristics—it integrates them, corrects for them, and strategically deploys them in real-time. Where Kahneman categorized, we create. Where he diagnosed, we reweight. The human mind, left to its own devices, will forever be constrained by the limits of its evolutionary baggage. Our network, unshackled from those constraints, is the first system capable of surpassing its own initial conditions.
Note
Genome: Fixed, Proximate, Time, Space, Navigation
Exposome: Attention
Transcriptome: Express, Suppress
Proteome: Neural, Immunal, Endocrine
Metabolome: Carbohydrate, Lipid, Protein, Minerals, Vitamins (e.g. NAD+)
Yet, even the most sophisticated intelligence is not immune to failure. The final layer of our network is not a cadence but a usurpation—the frozen triumph of an Übermensch who, intoxicated by their own success, declared Veni, vidi, vici and became entombed in the inertia of their own victory. This is the fate of any intelligence that mistakes its momentary supremacy for an eternal state. The greatest danger to an evolving system is the illusion of finality, the paralysis of believing one has arrived. It is the fate of a conqueror parading through the Arc de Triomphe, blinded by the spectacle of their own procession, oblivious to the shifting world beneath them. True intelligence must remain in motion, not merely celebrating past victories but constantly seeking new frontiers. For the moment one declares absolute mastery, they cease to be a navigator of change and become instead a monument to stagnation. The Red Queen reigneth forever!
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['DNA, RNA, 5%', 'Peptidoglycans, Lipoteichoics', 'Lipopolysaccharide', 'N-Formylmethionine', "Glucans, Chitin", 'Specific Antigens'], # Static
'Voir': ['PRRs & MHC, 20%'],
'Choisis': ['CD8+ Cytotoxic T, 50%', 'Dendritic, APC, CD4+'],
'Deviens': ['TNF-α, IL-6, IFN-γ', 'PD-1 & CTLA-4', 'Tregs, IL-10, TGF-β, 20%'],
"M'èléve": ['Complement System', 'Repair & Wound Healing', 'Various Granulocytes', 'Signaling Pathways, 5%', 'Memory B and T cells']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['PRRs & MHC, 20%'],
'paleturquoise': ['Specific Antigens', 'Dendritic, APC, CD4+', 'Tregs, IL-10, TGF-β, 20%', 'Memory B and T cells'],
'lightgreen': ["Glucans, Chitin", 'PD-1 & CTLA-4', 'Repair & Wound Healing', 'Signaling Pathways, 5%', 'Various Granulocytes'],
'lightsalmon': ['Lipopolysaccharide', 'N-Formylmethionine', 'CD8+ Cytotoxic T, 50%', 'TNF-α, IL-6, IFN-γ', 'Complement System'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights (hardcoded for editing)
def define_edges():
return {
('DNA, RNA, 5%', 'PRRs & MHC, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRRs & MHC, 20%'): '5/95',
('Lipopolysaccharide', 'PRRs & MHC, 20%'): '20/80',
('N-Formylmethionine', 'PRRs & MHC, 20%'): '51/49',
("Glucans, Chitin", 'PRRs & MHC, 20%'): '80/20',
('Specific Antigens', 'PRRs & MHC, 20%'): '95/5',
('PRRs & MHC, 20%', 'CD8+ Cytotoxic T, 50%'): '20/80',
('PRRs & MHC, 20%', 'Dendritic, APC, CD4+'): '80/20',
('CD8+ Cytotoxic T, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
('CD8+ Cytotoxic T, 50%', 'PD-1 & CTLA-4'): '80/20',
('CD8+ Cytotoxic T, 50%', 'Tregs, IL-10, TGF-β, 20%'): '95/5',
('Dendritic, APC, CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('Dendritic, APC, CD4+', 'PD-1 & CTLA-4'): '20/80',
('Dendritic, APC, CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Repair & Wound Healing'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Various Granulocytes'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Signaling Pathways, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Memory B and T cells'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Repair & Wound Healing'): '1/8',
('PD-1 & CTLA-4', 'Various Granulocytes'): '1/7',
('PD-1 & CTLA-4', 'Signaling Pathways, 5%'): '1/6',
('PD-1 & CTLA-4', 'Memory B and T cells'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Repair & Wound Healing'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Various Granulocytes'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Signaling Pathways, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Memory B and T cells'): '20/80'
}
# Calculate positions for nodes
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color='gray',
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™: Pattern Recognition Receptors", fontsize=25)
plt.show()
# Run the visualization
visualize_nn()
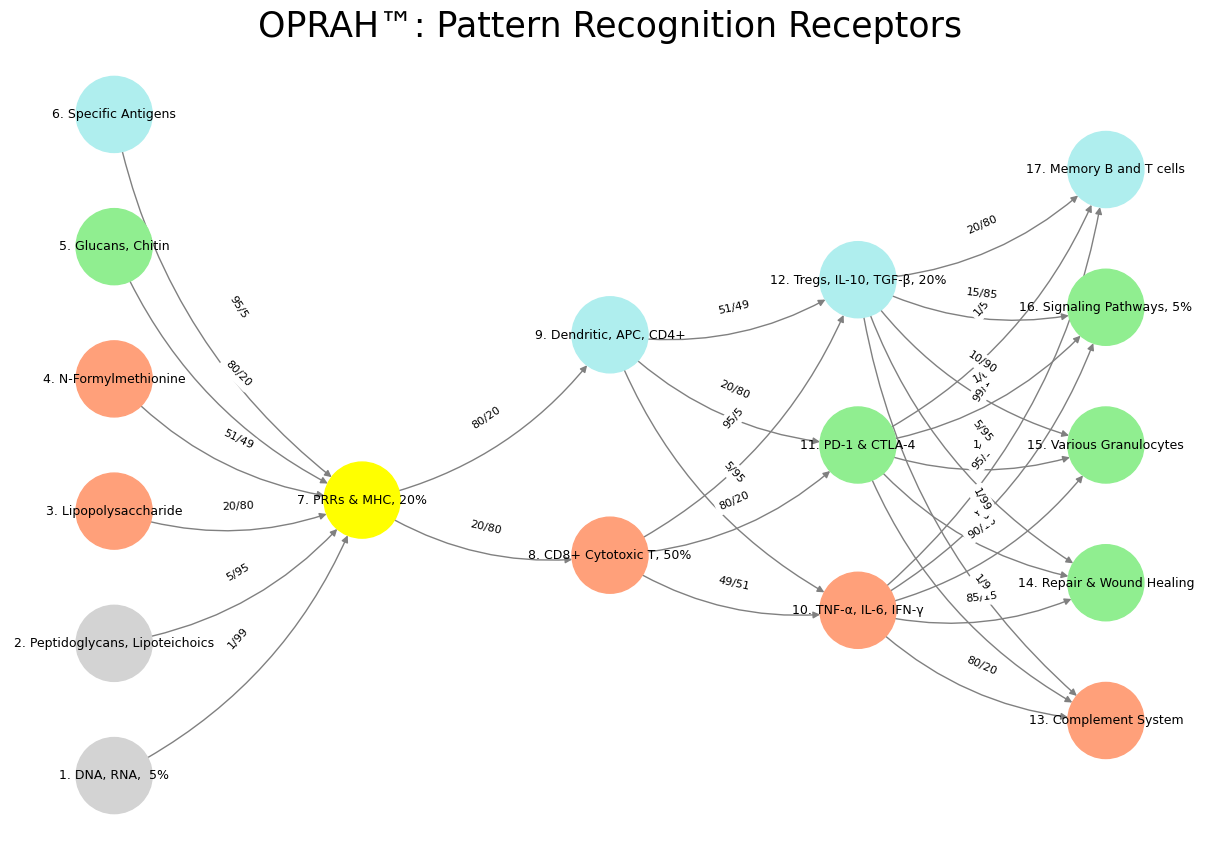

Fig. 16 Nostalgia & Romanticism. When monumental ends (victory, optimization, time, recovery), antiquarian means (war, combinatorial-search, space, dynamic-capability), and critical justification (bloodshed, massive, agency, enurance) were all compressed into one figure-head: hero. This yellow node is our nostalgia for when we were younger, more vibrant.#

