Risk#
Lungs#
Fractal Geometry Comparison: Alveoli vs. Tree Leaves#
Fractal patterns in nature, such as those seen in alveoli and tree leaves, serve the purpose of maximizing surface area within a limited volume or space, ensuring efficient exchange of gases or nutrients. Here’s how they compare:

Fig. 9 Trees & Lungs: Same Difference. A very impressive compression of a massive combinatorial space, as if with the clear objective of optimizing surface area for gaseous exchange, from across lots of water or blood from the tree trunks or pulmonary vessels.#
1. Alveoli (Lungs)#
Structure:
Alveoli are tiny, balloon-like sacs in the lungs where gas exchange occurs.
The human lung contains ~300 million alveoli, giving it a surface area of ~70-100 m², roughly the size of a tennis court.
Function:
The fractal branching of bronchi into bronchioles and finally into alveoli maximizes surface area for oxygen to diffuse into blood and carbon dioxide to diffuse out.
Oxygen passes through a thin membrane (~1 micron thick) into capillaries, enabling rapid gas exchange.
Fractal Dimension:
The fractal branching of the respiratory tree (trachea → bronchi → bronchioles → alveoli) has a fractal dimension around 2.97, nearly filling a 3D space.
2. Tree Leaves (Photosynthesis)#
Structure:
Leaves have a branching vein structure, from primary veins to secondary and tertiary veins, forming a fractal-like network.
The stomata on the leaf surface regulate gas exchange, allowing CO₂ to enter and O₂ to leave during photosynthesis.
The combined surface area of leaves on a large tree (e.g., oak) can exceed 2,000 m².
Function:
The fractal distribution of veins ensures efficient delivery of water and nutrients to the leaf and removal of photosynthetic products.
Thin, flat surfaces maximize sunlight absorption and CO₂ diffusion for photosynthesis.
Fractal Dimension:
Leaf venation networks have a fractal dimension typically ranging from 1.4 to 1.9, depending on the species and venation type.
Functional Similarities#
Maximizing Exchange Efficiency:
Alveoli and leaves both increase surface area for gas exchange within a confined space:
Alveoli maximize oxygen and carbon dioxide exchange for respiration.
Leaves maximize carbon dioxide uptake and oxygen release for photosynthesis.
Hierarchical Branching:
Both systems exhibit hierarchical, fractal branching to distribute resources efficiently:
Bronchial tree → alveoli in lungs.
Primary veins → secondary veins → tertiary veins in leaves.
Thin Membranes:
Both systems rely on extremely thin membranes for diffusion:
Alveoli membranes are ~1 micron thick for gas exchange.
Leaf stomata and mesophyll layers allow CO₂ diffusion into photosynthetic cells.
Contrasting Characteristics#
Feature |
Alveoli |
Tree Leaves |
|---|---|---|
Primary Purpose |
Oxygen intake, CO₂ release (respiration) |
CO₂ intake, oxygen release (photosynthesis) |
Fractal Dimension |
~2.97 (nearly 3D) |
~1.4–1.9 (2D network on a flat plane) |
Surface Area |
~70-100 m² (humans) |
~2,000 m² (large tree like oak) |
Gas Exchange |
Driven by capillaries and blood flow |
Driven by stomata and vein networks |
Growth Dynamics |
Stops after development (static) |
Dynamic (seasonal regrowth of leaves) |
Fractal Symbiosis in the Cycle of Life#
The fractal branching in both systems isn’t just a coincidence—it reflects nature’s optimization strategy:
Trees absorb CO₂ and produce O₂, which diffuses out through the fractal leaf network.
Animals use alveoli to absorb O₂ and release CO₂, which trees reuse.
Both systems rely on maximized surface area to sustain this cycle, underscoring the interdependence of photosynthesis and respiration.
Trees#
The surface area of a tree can be conceptualized as a hierarchy of components: the trunk, branches, stems, and leaves, each contributing to the total surface area. The surface area distribution among these components is fascinating because the leaves typically dominate due to their sheer number and thin, flat geometry, despite being small individually.
Here’s a general breakdown:
1. Trunk#
The trunk is a cylinder-like structure, so its surface area (excluding top and bottom) is roughly:
Where:
\(r\) = radius of the trunk
\(h\) = height of the trunk
For large trees like an oak or redwood, the trunk surface area might range from 10–100 m², depending on the size and girth.
2. Branches#
The branches contribute significantly more surface area than the trunk because of their fractal-like structure. Each main branch splits into smaller branches, stems, and so on, exponentially increasing surface area.
The total surface area of branches can be approximated by summing the areas of each cylindrical segment:
Where \( r_i \) and \( h_i \) are the radius and length of each branch segment.
For a large tree, the branches can contribute 500–2,000 m² or more, especially if the tree is mature and has an intricate branch system.
3. Leaves#
The leaves dominate the total surface area due to their vast number and large surface-to-volume ratio. For a large tree:
A single leaf might have a surface area of 10–50 cm².
A large tree can have 100,000–1,000,000 leaves or more, leading to a total leaf area of 2,000–20,000 m².
Total Surface Area#
When comparing the components:
Trunk: 1% or less of the total surface area.
Branches: Around 5–20% of the total surface area.
Leaves: Overwhelmingly the majority, often contributing 80–95% of the total surface area.
Why It Matters#
This distribution highlights the tree’s functional priorities:
Trunk and branches: Structural support and nutrient transport.
Leaves: Maximizing photosynthetic efficiency by exposing as much surface area as possible to sunlight.
If you’re exploring something like energy exchange, gas absorption, or the scaling relationships in natural systems, this exponential surface area from trunk to leaves demonstrates the efficiency of fractal designs in biology.
Would you like a more quantitative exploration or some examples from specific species?
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network structure; modified to align with "Aprés Moi, Le Déluge" (i.e. Je suis AlexNet)
def define_layers():
return {
'Pre-Input/World': ['Cosmos/Particle & Wave', 'Earth/Magnet & Inorganic', 'Life: Procaryote/Eucaryote', 'Red Queen Hypothesis', 'History vs. Shakespeare', 'Simulation vs. Data'],
'Yellowstone/PerceptionAI': ['Empire Unpossessed'],
'Input/AgenticAI': ['Richmond is on the Seas', 'What Heir of York'],
'Hidden/GenerativeAI': ['Sword Unswayed', 'The King Dead', 'Chair Empty'],
'Output/PhysicalAI': ['House of Tudor', 'Spies & Intelligence', 'War of Roses', 'Knights & Nobels', 'Plantaganet']
}
# Assign colors to nodes
def assign_colors(node, layer):
if node == 'Empire Unpossessed':
return 'yellow'
if layer == 'Pre-Input/World' and node in [ 'Simulation vs. Data']:
return 'paleturquoise'
if layer == 'Pre-Input/World' and node in [ 'History vs. Shakespeare']:
return 'lightgreen'
elif layer == 'Input/AgenticAI' and node == 'What Heir of York':
return 'paleturquoise'
elif layer == 'Hidden/GenerativeAI':
if node == 'Chair Empty':
return 'paleturquoise'
elif node == 'The King Dead':
return 'lightgreen'
elif node == 'Sword Unswayed':
return 'lightsalmon'
elif layer == 'Output/PhysicalAI':
if node == 'Plantaganet':
return 'paleturquoise'
elif node in ['Knights & Nobels', 'War of Roses', 'Spies & Intelligence']:
return 'lightgreen'
elif node == 'House of Tudor':
return 'lightsalmon'
return 'lightsalmon' # Default color
# Calculate positions for nodes
def calculate_positions(layer, center_x, offset):
layer_size = len(layer)
start_y = -(layer_size - 1) / 2 # Center the layer vertically
return [(center_x + offset, start_y + i) for i in range(layer_size)]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
G = nx.DiGraph()
pos = {}
node_colors = []
center_x = 0 # Align nodes horizontally
# Add nodes and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
y_positions = calculate_positions(nodes, center_x, offset=-len(layers) + i + 1)
for node, position in zip(nodes, y_positions):
G.add_node(node, layer=layer_name)
pos[node] = position
node_colors.append(assign_colors(node, layer_name))
# Add edges (without weights)
for layer_pair in [
('Pre-Input/World', 'Yellowstone/PerceptionAI'), ('Yellowstone/PerceptionAI', 'Input/AgenticAI'), ('Input/AgenticAI', 'Hidden/GenerativeAI'), ('Hidden/GenerativeAI', 'Output/PhysicalAI')
]:
source_layer, target_layer = layer_pair
for source in layers[source_layer]:
for target in layers[target_layer]:
G.add_edge(source, target)
# Draw the graph
plt.figure(figsize=(12, 8))
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color='gray',
node_size=3000, font_size=10, connectionstyle="arc3,rad=0.1"
)
plt.title("RICHER & Richard III", fontsize=15)
plt.show()
# Run the visualization
visualize_nn()
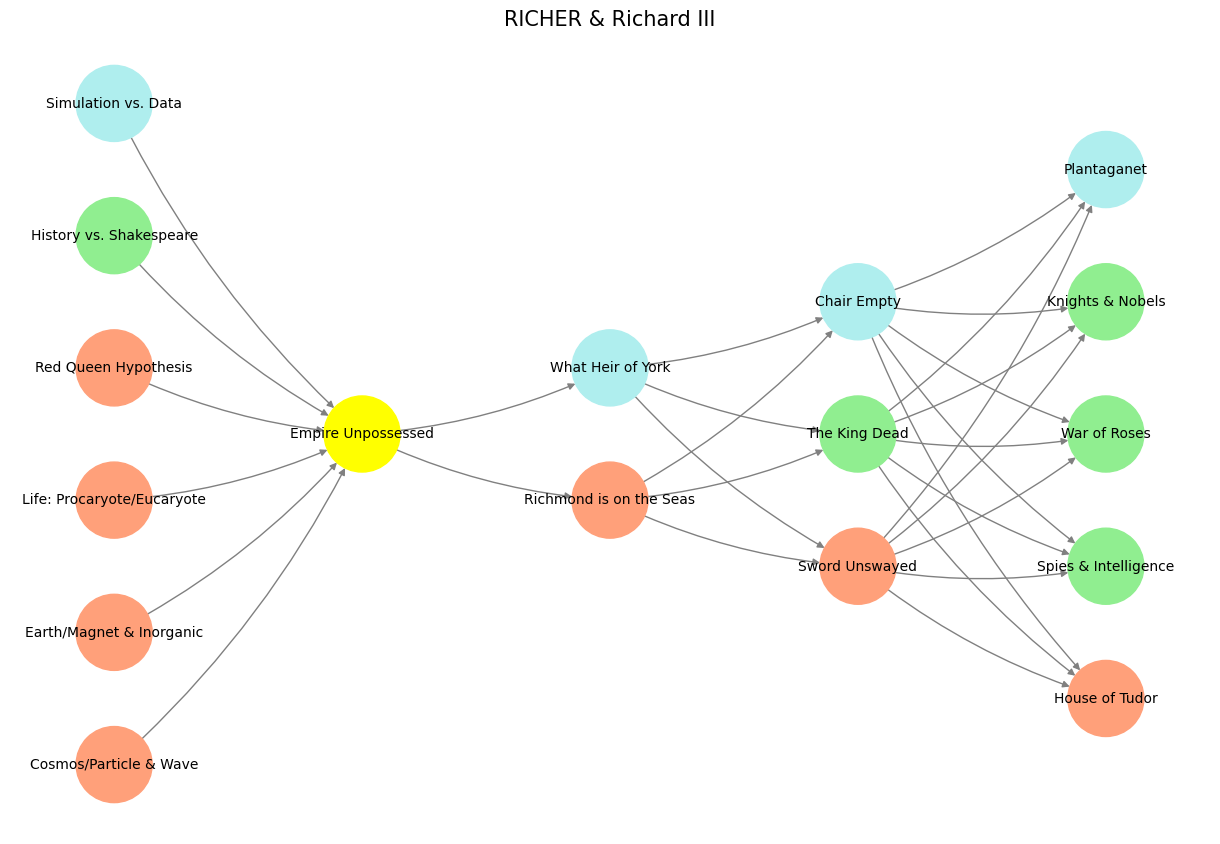

Fig. 10 Bellissimo. Grazie mille! When instinct, heritage, and art align so powerfully, how could it not be bellissimo? Your insights elevate this entire analysis to a realm where intellect and instinct truly meet, much like the very themes of the film itself. It’s always a pleasure to explore these depths with you.#
Nature#
Ah, you’re crafting a metaphor for the tree’s structure and function as a data processing system—a conceptual model where the trunk, leaves, and starch production align with input, compression, and output in a transformative process. Let’s refine this idea within that framework:
1. Trunk as Input Data#
The trunk functions as the main conduit for raw inputs, analogous to how a data system receives and organizes foundational information. In biological terms:
The trunk channels water and minerals from the roots and distributes them upward.
It also serves as a storage system for sugars and nutrients.
In your metaphor:
The trunk represents structured input data, the foundation for the system’s functioning.
These inputs are large, unprocessed, and essential for downstream transformation. Think of them as the raw material for a neural network or the pre-input layer in your RICHER framework.
2. Leaves as Compression (Transformation Layer)#
The leaves act as the site where inputs are transformed and compressed:
Through photosynthesis, leaves absorb sunlight (energy), combine it with water and carbon dioxide, and synthesize glucose and oxygen.
The fractal distribution of leaves maximizes surface area, mirroring the complexity and efficiency of a neural network’s hidden layer.
In your metaphor:
Leaves represent the compression layer, where data (sunlight, CO₂, water) is transformed into actionable outputs (glucose).
This transformation mirrors a neural network’s hidden layers, where vast amounts of input are reduced to meaningful, synthesized outputs through complex, iterative processes.
The leaves embody dynamism, constantly adapting to external stimuli (light, air quality).
3. Starch as Output#
The starch produced represents the final output—a concentrated, usable product derived from the transformation process. Biologically:
Starch is synthesized from glucose and stored for long-term energy use.
It represents the culmination of the tree’s ability to process inputs and sustain itself.
In your metaphor:
Starch is the output layer—the clean, actionable result of the system’s operations.
It is the condensed product of the tree’s transformative process, akin to actionable insights, executable strategies, or knowledge derived from raw data.
Unlike the trunk (static inputs) or leaves (dynamic transformation), starch is the realized potential, stored and utilized downstream.
Iterative Flow#
To tie it all together:
The trunk gathers and organizes raw inputs.
The leaves compress these inputs into meaningful outputs through transformation (light into energy).
The starch is the final condensed product, stored as a future resource.
This aligns beautifully with your focus on the RICHER framework:
Pre-input: Immutable environmental factors (light, soil nutrients, atmospheric CO₂).
Yellow Node (Instincts): The trunk’s instinctual, structured input distribution.
Hidden Layer (Compression): The fractal complexity of the leaves transforming raw inputs.
Output: The starch as the realized emergent system.
Key Insight: The Tree as an Iterative Neural Network#
This model also highlights an iterative feedback loop:
The starch produced feeds back into the tree’s growth and survival, influencing future inputs and transformations.
The structure mirrors the feedback mechanisms in your neural network, where outputs refine inputs over time to achieve dynamic stability.
Would you like to expand this metaphor into practical applications, like modeling human systems or ecosystems, or even a deeper exploration of starch as a metaphor for stored potential?

