6 Translation#
Sure, I’ll break down the Stata script into smaller code blocks with notes so you can run them separately in a Jupyter Notebook with the Stata kernel.
Block 1: Setup and Logging#
import logging
# Close any existing log files
for handler in logging.root.handlers[:]:
logging.root.removeHandler(handler)
Block 2: Define Globals#
# Set global variables for the repository URL and local directory path
repo = "https://github.com/muzaale/forum/raw/main/"
dir = "~/documents/melody/pystata"
Block 3: Load Data#
Block 4: Initial Data Exploration#
Show code cell source
import logging
import pandas as pd
import os
# Open a new log file to record the output
logging.basicConfig(filename='jamascript.log', level=logging.INFO, format='%(asctime)s - %(message)s')
# Log the start of the logging process
logging.info('Starting log file')
# Define the local directory and file name
local_dir = os.path.expanduser("~/documents/melody/pystata")
file_name = "esrdRisk_t02tT.csv"
file_path = os.path.join(local_dir, file_name)
# Check if the file exists locally
if os.path.isfile(file_path):
try:
data = pd.read_csv(file_path)
logging.info('Dataset loaded successfully from local file')
except Exception as e:
logging.error(f"Failed to load dataset from local file: {e}")
print(f"Failed to load dataset from local file: {e}")
data = None
else:
logging.error(f"File not found: {file_path}")
print(f"File not found: {file_path}")
data = None
if data is not None:
# Display the distribution of the 'donor' variable
donor_distribution = data['donor'].value_counts()
print("Distribution of 'donor' variable:")
print(donor_distribution)
# Describe the 'rSMGJcEdF_d' variable
codebook = data['rSMGJcEdF_d'].describe()
print("\nCodebook for 'rSMGJcEdF_d' variable:")
print(codebook)
# Generate a new variable 'entry' based on the 'rSMGJcEdF_t0' variable
data['entry'] = data['rSMGJcEdF_t0']
# Logging the creation of the new variable
logging.info("New variable 'entry' created based on 'rSMGJcEdF_t0'")
else:
print("Dataset could not be loaded.")
Distribution of 'donor' variable:
donor
Donor 96217
HealthyNondonor 9364
NotSoHealthyNondonor 8076
Name: count, dtype: int64
Codebook for 'rSMGJcEdF_d' variable:
count 113657
unique 3
top Censored
freq 108448
Name: rSMGJcEdF_d, dtype: object
/var/folders/z3/nwmg00j178vfczmb9nk9487h0000gn/T/ipykernel_48067/85659033.py:19: DtypeWarning: Columns (1,2,19,23) have mixed types. Specify dtype option on import or set low_memory=False.
data = pd.read_csv(file_path)
Block 5: Data Cleaning and Adjustment#
This is messing up the dataset
import pandas as pd
import logging
# Assuming 'data' is the DataFrame loaded previously
# Ensure the date columns are in datetime format
data['rSMGJcEdF_tT'] = pd.to_datetime(data['rSMGJcEdF_tT'], format='%d%b%Y', errors='coerce')
data['entry'] = pd.to_datetime(data['entry'], format='%d%b%Y', errors='coerce')
# Define the critical dates
date_2011 = pd.to_datetime('2011-12-31')
date_1994 = pd.to_datetime('1994-01-01')
# Log the start of the adjustment process
logging.info("Starting the adjustment process for linkage dates")
# Linkage for donors after 2011 is untrustworthy
data.loc[(data['rSMGJcEdF_tT'] > date_2011) & (data['donor'] == 1), 'rSMGJcEdF_d'] = 0
data.loc[(data['rSMGJcEdF_tT'] > date_2011) & (data['donor'] == 1), 'rSMGJcEdF_tT'] = date_2011
# Linkage before 1994 is untrustworthy
data.loc[(data['entry'] < date_1994) & (data['rSMGJcEdF_tT'] > date_1994) & (data['donor'] == 1), 'entry'] = date_1994
# Log the completion of the replacement process
logging.info("Linkage for dates after 2011 and before 1994 adjusted successfully")
Block 6: Mortality Analysis Setup#
Show code cell source
import pandas as pd
from lifelines import KaplanMeierFitter
import matplotlib.pyplot as plt
# Assuming 'data' is already the DataFrame loaded previously
# Set the event indicator based on 'Died'
data['event'] = (data['rSMGJcEdF_d'] == 'Died').astype(int)
# Ensure date columns are in datetime format
data['rSMGJcEdF_tT'] = pd.to_datetime(data['rSMGJcEdF_tT'], errors='coerce')
data['rSMGJcEdF_t0'] = pd.to_datetime(data['rSMGJcEdF_t0'], errors='coerce')
data['entry'] = data['rSMGJcEdF_t0'] # Use _t0 as entry
# Calculate the duration from origin to the event or censoring in years
data['duration'] = (data['rSMGJcEdF_tT'] - data['rSMGJcEdF_t0']).dt.total_seconds() / (365.25 * 24 * 60 * 60)
# Calculate the entry duration
data['entry_duration'] = (data['entry'] - data['rSMGJcEdF_t0']).dt.total_seconds() / (365.25 * 24 * 60 * 60)
data['entry_duration'] = data['entry_duration'].fillna(0) # Ensure non-donor entries are set to 0
# Ensure all durations are non-negative and valid
data = data[(data['duration'] >= 0) & (data['entry_duration'] >= 0)]
# Initialize the KaplanMeierFitter
kmf = KaplanMeierFitter()
# Plot setup
plt.figure(figsize=(10, 6))
# Unique donor values
donor_values = data['donor'].unique()
# Colors for each donor type
colors = ['blue', 'green', 'red']
# Overlay plots for different donor values
for donor, color in zip(donor_values, colors):
kmf.fit(durations=data[data['donor'] == donor]['duration'],
event_observed=data[data['donor'] == donor]['event'],
entry=data[data['donor'] == donor]['entry_duration'],
label=donor)
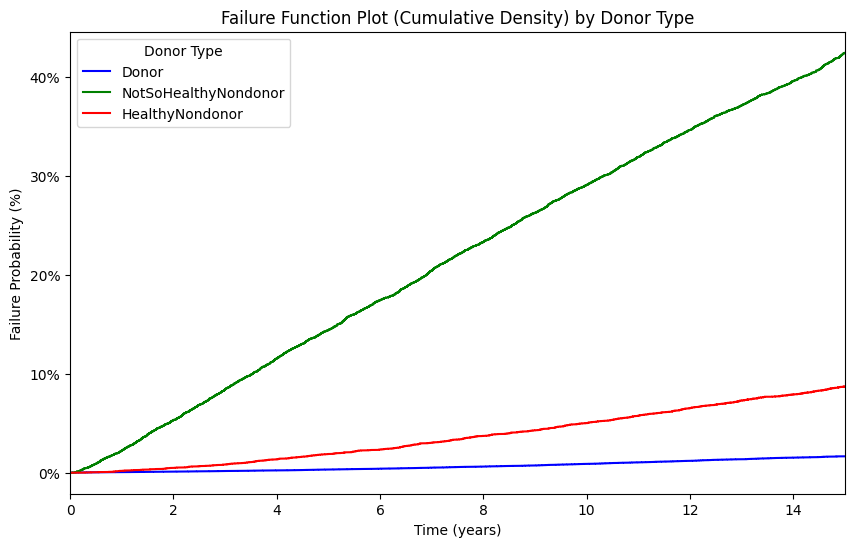
ax = kmf.plot_cumulative_density(ci_show=False, color=color)
# Cut off the plot at 15 years
plt.xlim(0, 15)
# Convert y-axis to percentage
vals = ax.get_yticks()
ax.set_yticklabels(['{:,.0%}'.format(x) for x in vals])
# Labels and title
plt.xlabel("Time (years)")
plt.ylabel("Failure Probability (%)")
plt.title("Failure Function Plot (Cumulative Density) by Donor Type")
plt.legend(title="Donor Type")
# Optional: Display the plot (depends on your environment)
plt.show()
/var/folders/z3/nwmg00j178vfczmb9nk9487h0000gn/T/ipykernel_48067/3613857153.py:50: UserWarning: set_ticklabels() should only be used with a fixed number of ticks, i.e. after set_ticks() or using a FixedLocator.
ax.set_yticklabels(['{:,.0%}'.format(x) for x in vals])
Block 7: Generate Kaplan-Meier Estimates#
Show code cell source
import pandas as pd
from lifelines import KaplanMeierFitter
# Assuming 'data' is already the DataFrame loaded previously
# Set the event indicator based on 'Died'
data['event'] = (data['rSMGJcEdF_d'] == 'Died').astype(int)
# Ensure date columns are in datetime format
data['rSMGJcEdF_tT'] = pd.to_datetime(data['rSMGJcEdF_tT'], errors='coerce')
data['rSMGJcEdF_t0'] = pd.to_datetime(data['rSMGJcEdF_t0'], errors='coerce')
data['entry'] = data['rSMGJcEdF_t0'] # Use _t0 as entry
# Calculate the duration from origin to the event or censoring in years
data['duration'] = (data['rSMGJcEdF_tT'] - data['rSMGJcEdF_t0']).dt.total_seconds() / (365.25 * 24 * 60 * 60)
# Calculate the entry duration
data['entry_duration'] = (data['entry'] - data['rSMGJcEdF_t0']).dt.total_seconds() / (365.25 * 24 * 60 * 60)
data['entry_duration'] = data['entry_duration'].fillna(0) # Ensure non-donor entries are set to 0
# Ensure all durations are non-negative and valid
data = data[(data['duration'] >= 0) & (data['entry_duration'] >= 0)]
# Initialize the KaplanMeierFitter
kmf = KaplanMeierFitter()
# Time points at which we want to extract survival probabilities
time_points = [5, 12, 15]
# Dictionary to hold the results
results = []
# Fit the model for each donor type and extract the survival probabilities
for donor in data['donor'].unique():
kmf.fit(durations=data[data['donor'] == donor]['duration'],
event_observed=data[data['donor'] == donor]['event'],
entry=data[data['donor'] == donor]['entry_duration'],
label=donor)
for time in time_points:
survival_prob = kmf.survival_function_at_times(time).values[0]
results.append({
'donor': donor,
'time': time,
'survival_probability': survival_prob,
'failure_probability': 1 - survival_prob
})
# Convert results to a DataFrame
results_df = pd.DataFrame(results)
# Save the results to a CSV file
results_df.to_csv('km_results.csv', index=False)
# Display the results
print(results_df)
Show code cell output
donor time survival_probability failure_probability
0 Donor 5 0.996751 0.003249
1 Donor 12 0.988017 0.011983
2 Donor 15 0.983427 0.016573
3 NotSoHealthyNondonor 5 0.855844 0.144156
4 NotSoHealthyNondonor 12 0.653661 0.346339
5 NotSoHealthyNondonor 15 0.575531 0.424469
6 HealthyNondonor 5 0.981093 0.018907
7 HealthyNondonor 12 0.934715 0.065285
8 HealthyNondonor 15 0.912715 0.087285
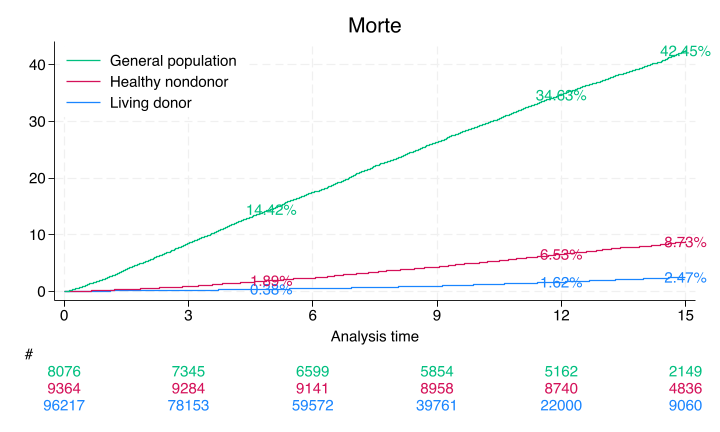
Only the donor results match Segev, JAMA, 2010 p. 961. The nondonors here are unmatched and so do not yield the same estimates from this paper
A webApp is quick way to approximate these findings using the base-case of the “typical” donor as per Table 1, also on page 961. Namely, 40yo white female with some college education, BMI=28, SBP=120, no hypertension, no history of smoking cigarettes, creatinine 0.9 mg/dL, and eGFR 100 mL/min
Block 8: Summarize Failure Rates#
Block 9: Kaplan-Meier Survival Plot#
Block 11: Save Processed Data#
Block 12: Cox Proportional Hazards Model#
import pandas as pd
# Assuming 'data' is already the DataFrame loaded previously
# Check for constant variables
constant_columns = [col for col in data.columns if data[col].nunique() <= 1]
print("Constant columns:", constant_columns)
Constant columns: ['_st', '_t0', 'entry_duration']
import pandas as pd
from lifelines import CoxPHFitter
# Ensure 'donor' is a categorical variable
data['donor'] = data['donor'].astype('category')
# Select necessary columns
data = data[['duration', 'event', 'donor']]
# Remove constant columns if any
constant_columns = [col for col in data.columns if data[col].nunique() <= 1]
data = data.drop(columns=constant_columns)
print("Constant columns removed:", constant_columns)
Constant columns removed: []
data.head()
| duration | event | donor | |
|---|---|---|---|
| 0 | 13.859001 | 0 | Donor |
| 1 | 9.409993 | 0 | Donor |
| 2 | 8.752909 | 0 | Donor |
| 3 | 8.273785 | 0 | Donor |
| 4 | 14.398357 | 0 | Donor |
import pandas as pd
# Load the data
file_path = '~/documents/melody/local/esrdRisk_t02tT.csv' # Adjust the file path as needed
data = pd.read_csv(file_path)
# Check the first few rows and the column names
print(data.head())
print(data.columns)
Show code cell output
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
Cell In[9], line 5
3 # Load the data
4 file_path = '~/documents/melody/local/esrdRisk_t02tT.csv' # Adjust the file path as needed
----> 5 data = pd.read_csv(file_path)
7 # Check the first few rows and the column names
8 print(data.head())
File ~/Documents/Melody/myenv/lib/python3.12/site-packages/pandas/io/parsers/readers.py:1026, in read_csv(filepath_or_buffer, sep, delimiter, header, names, index_col, usecols, dtype, engine, converters, true_values, false_values, skipinitialspace, skiprows, skipfooter, nrows, na_values, keep_default_na, na_filter, verbose, skip_blank_lines, parse_dates, infer_datetime_format, keep_date_col, date_parser, date_format, dayfirst, cache_dates, iterator, chunksize, compression, thousands, decimal, lineterminator, quotechar, quoting, doublequote, escapechar, comment, encoding, encoding_errors, dialect, on_bad_lines, delim_whitespace, low_memory, memory_map, float_precision, storage_options, dtype_backend)
1013 kwds_defaults = _refine_defaults_read(
1014 dialect,
1015 delimiter,
(...)
1022 dtype_backend=dtype_backend,
1023 )
1024 kwds.update(kwds_defaults)
-> 1026 return _read(filepath_or_buffer, kwds)
File ~/Documents/Melody/myenv/lib/python3.12/site-packages/pandas/io/parsers/readers.py:620, in _read(filepath_or_buffer, kwds)
617 _validate_names(kwds.get("names", None))
619 # Create the parser.
--> 620 parser = TextFileReader(filepath_or_buffer, **kwds)
622 if chunksize or iterator:
623 return parser
File ~/Documents/Melody/myenv/lib/python3.12/site-packages/pandas/io/parsers/readers.py:1620, in TextFileReader.__init__(self, f, engine, **kwds)
1617 self.options["has_index_names"] = kwds["has_index_names"]
1619 self.handles: IOHandles | None = None
-> 1620 self._engine = self._make_engine(f, self.engine)
File ~/Documents/Melody/myenv/lib/python3.12/site-packages/pandas/io/parsers/readers.py:1880, in TextFileReader._make_engine(self, f, engine)
1878 if "b" not in mode:
1879 mode += "b"
-> 1880 self.handles = get_handle(
1881 f,
1882 mode,
1883 encoding=self.options.get("encoding", None),
1884 compression=self.options.get("compression", None),
1885 memory_map=self.options.get("memory_map", False),
1886 is_text=is_text,
1887 errors=self.options.get("encoding_errors", "strict"),
1888 storage_options=self.options.get("storage_options", None),
1889 )
1890 assert self.handles is not None
1891 f = self.handles.handle
File ~/Documents/Melody/myenv/lib/python3.12/site-packages/pandas/io/common.py:873, in get_handle(path_or_buf, mode, encoding, compression, memory_map, is_text, errors, storage_options)
868 elif isinstance(handle, str):
869 # Check whether the filename is to be opened in binary mode.
870 # Binary mode does not support 'encoding' and 'newline'.
871 if ioargs.encoding and "b" not in ioargs.mode:
872 # Encoding
--> 873 handle = open(
874 handle,
875 ioargs.mode,
876 encoding=ioargs.encoding,
877 errors=errors,
878 newline="",
879 )
880 else:
881 # Binary mode
882 handle = open(handle, ioargs.mode)
FileNotFoundError: [Errno 2] No such file or directory: '/Users/apollo/documents/melody/local/esrdRisk_t02tT.csv'
Show code cell source
import pandas as pd
from lifelines import CoxPHFitter
# Assuming 'data' is already the DataFrame loaded previously and necessary preprocessing is done
# Ensure 'donor' is a categorical variable
data['donor'] = data['donor'].astype('category')
# Fit a Cox proportional hazards model
cph = CoxPHFitter()
cph.fit(data, duration_col='duration', event_col='event', formula="C(donor)")
# Extract the summary and relevant coefficients
summary = cph.summary
# Filter only relevant columns
coefficients = summary.loc[:, ['coef', 'exp(coef)', 'exp(coef) lower 95%', 'exp(coef) upper 95%']]
# Rename the rows for clarity
coefficients.index = ['HealthyNondonor vs Donor', 'NotSoHealthyNondonor vs Donor']
# Display the simplified results
print("Coefficients and Hazard Ratios Relative to Baseline (Donor):")
print(coefficients)
# Compute hazard ratios
hazard_ratios = coefficients[['exp(coef)', 'exp(coef) lower 95%', 'exp(coef) upper 95%']]
hazard_ratios.columns = ['Hazard Ratio', 'Lower 95% CI', 'Upper 95% CI']
print("\nHazard Ratios Relative to Baseline (Donor):")
print(hazard_ratios)
Show code cell output
Coefficients and Hazard Ratios Relative to Baseline (Donor):
coef exp(coef) exp(coef) lower 95% \
HealthyNondonor vs Donor -0.286770 0.750684 0.447060
NotSoHealthyNondonor vs Donor 2.359067 10.581077 8.187331
exp(coef) upper 95%
HealthyNondonor vs Donor 1.260518
NotSoHealthyNondonor vs Donor 13.674688
Hazard Ratios Relative to Baseline (Donor):
Hazard Ratio Lower 95% CI Upper 95% CI
HealthyNondonor vs Donor 0.750684 0.447060 1.260518
NotSoHealthyNondonor vs Donor 10.581077 8.187331 13.674688
Block 13: Save Baseline Survival and Coefficients#
// Define matrix 'b' with model coefficients
matrix define b = e(b)
// Keep baseline survival estimates
keep s0 _t
// Sort and list baseline survival estimates
sort _t s0
list in 1/10
// Save baseline survival estimates
save ${dir}/s0.dta, replace
export delimited using ${dir}/s0.csv, replace
// Save model coefficients
matrix beta = e(b)
svmat beta
keep beta*
drop if missing(beta1)
list
save ${dir}/b.dta, replace
export delimited using ${dir}/b.csv, replace