Traditional#
Here’s something to chew on:
Great works of art endure because they interrogate dysfunction, particularly at the relational level—marriage, family, community, tribe, city, and ideology. They do so because dysfunction is an existential threat. A core principle of immunology is that the body must recognize non-self as soon as possible and eliminate it, lest dysfunction cascade into systemic failure. But in art, as in biology, the threshold of recognition is fraught with complications. The immune system’s sensitivity must balance precision with tolerance; too lax, and threats infiltrate unchecked; too aggressive, and it destroys its own host. The same dilemma plays out in literature, drama, and cinema. Tragedy, nihilism, and comedy are artistic explorations of this delicate equilibrium, where misrecognition—whether too early, too late, or too biased—determines fate.

Fig. 36 I’d advise you to consider your position carefully (layer 3 fork in the road), perhaps adopting a more flexible posture (layer 4 dynamic capabilities realized), while keeping your ear to the ground (layer 2 yellow node), covering your retreat (layer 5 Athena’s shield, helmet, and horse), and watching your rear (layer 1 ecosystem and perspective).#
Tragedy reveals what happens when dysfunction is recognized either too late or when the response to it is maladaptive. In King Lear, Lear fails to recognize which of his daughters truly love him. His error sets in motion a cascade of destruction, much like a failing immune system that mistakes friend for foe and unleashes an autoimmune storm. The same principle applies to Othello, where Iago manipulates perception, altering Othello’s threshold for recognizing threat. Othello, like an immune system turned against its own cells, destroys Desdemona, ultimately self-destructing in the process. The lesson of tragedy is clear: misjudging non-self—either failing to detect it in time or mistaking self for non-self—is fatal.
Nihilism emerges when dysfunction is recognized, but no viable resolution exists. When all equilibria collapse, when every relational structure—marriage, family, community—fails, what remains is despair or absurdity. This is the core tension in Hamlet’s existential paralysis. He perceives the rot in Denmark, but his attempts at resolution—whether through feigned madness, inaction, or violent vengeance—fail to restore equilibrium. The immune system has no answer to the corruption; it either overreacts or withers into inaction, leaving the host in a liminal space between life and death. The modern nihilistic response to dysfunction is resignation, the belief that the system is beyond repair. But this too is an adaptive failure, one that great art forces us to confront.
If tragedy and nihilism focus on failure, early recognition is the province of the strategic figure. These characters detect dysfunction before others do and act preemptively. Shakespeare’s Henry V is the archetypal strategic immune cell, recognizing threats and recalibrating alliances before they metastasize. He eliminates internal threats (Falstaff, his former self) and external ones (France) with a ruthless efficiency that ensures survival. This is the immune system at its most disciplined, eliminating precancerous lesions before they become tumors. But this comes at a cost—his relational bonds are sacrificed for the sake of the body politic. The same is true of Machiavellian figures like Richard III, who, though doomed in the end, exemplify early recognition taken to its extreme.
Yet not all responses to dysfunction must be adversarial. Art also probes how relational equilibria—cooperative, transactional, or adversarial—dictate outcomes. Shakespeare’s comedies, for instance, test whether dysfunction can be resolved without destruction. Much Ado About Nothing and Twelfth Night show relationships on the verge of collapse, only to be saved by wit, disguise, and misdirection. The immune system, too, has a regulatory layer. It does not merely attack; it negotiates, forming tolerogenic responses that allow for co-existence rather than annihilation. In art, this is the domain of comedy—the space where dysfunction is acknowledged but repurposed into new forms of relational stability.
Absurdity and humor arise when the noise-to-signal ratio is so high that pattern recognition fails. This is why the fool, the joker, and the trickster play such an integral role in art. They expose the arbitrary nature of recognition itself. In Waiting for Godot, the characters wait for meaning, for structure, for the recognition of an immune signal that never comes. But the joke is that life is precisely this—an immune system scanning endlessly, detecting nothing, reacting to phantoms, and mistaking signals for noise. If King Lear is immunological tragedy—where recognition fails—Godot is immunological comedy, where recognition is forever deferred.
All great art wrestles with the fundamental question: how do we recognize dysfunction, and what do we do about it? Too much sensitivity, and we destroy ourselves through paranoia, prejudice, and overreaction. Too little, and dysfunction consumes the system. The balance is never stable, never absolute. Shakespeare understood this in The Tempest, where Prospero calibrates his recognition of threats over time, shifting from vengeance to reconciliation. Immunology, like great storytelling, is about managing thresholds of perception, knowing when to attack, when to tolerate, and when to transform. Art matters because it trains us in this recognition, forcing us to confront our own biases, our own thresholds of tolerance and intolerance, and ultimately, our own survival.
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['DNA, RNA, 5%', 'Peptidoglycans, Lipoteichoics', 'Lipopolysaccharide', 'N-Formylmethionine', "Glucans, Chitin", 'Specific Antigens'],
'Voir': ['PRR & ILCs, 20%'],
'Choisis': ['CD8+, 50%', 'CD4+'],
'Deviens': ['TNF-α, IL-6, IFN-γ', 'PD-1 & CTLA-4', 'Tregs, IL-10, TGF-β, 20%'],
"M'èléve": ['Complement System', 'Platelet System', 'Granulocyte System', 'Innate Lymphoid Cells, 5%', 'Adaptive Lymphoid Cells']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['PRR & ILCs, 20%'],
'paleturquoise': ['Specific Antigens', 'CD4+', 'Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'],
'lightgreen': ["Glucans, Chitin", 'PD-1 & CTLA-4', 'Platelet System', 'Innate Lymphoid Cells, 5%', 'Granulocyte System'],
'lightsalmon': ['Lipopolysaccharide', 'N-Formylmethionine', 'CD8+, 50%', 'TNF-α, IL-6, IFN-γ', 'Complement System'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights
def define_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
('PRR & ILCs, 20%', 'CD8+, 50%'): '20/80',
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD8+, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
('CD8+, 50%', 'PD-1 & CTLA-4'): '80/20',
('CD8+, 50%', 'Tregs, IL-10, TGF-β, 20%'): '95/5',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD4+', 'PD-1 & CTLA-4'): '20/80',
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Platelet System'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Granulocyte System'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Innate Lymphoid Cells, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Adaptive Lymphoid Cells'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Platelet System'): '1/8',
('PD-1 & CTLA-4', 'Granulocyte System'): '1/7',
('PD-1 & CTLA-4', 'Innate Lymphoid Cells, 5%'): '1/6',
('PD-1 & CTLA-4', 'Adaptive Lymphoid Cells'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Platelet System'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Granulocyte System'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Define edges to be highlighted in black
def define_black_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
}
# Calculate node positions
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
black_edges = define_black_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
edge_colors = []
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
edge_colors.append('black' if (source, target) in black_edges else 'lightgrey')
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color=edge_colors,
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™ aAPCs", fontsize=18)
plt.show()
# Run the visualization
visualize_nn()
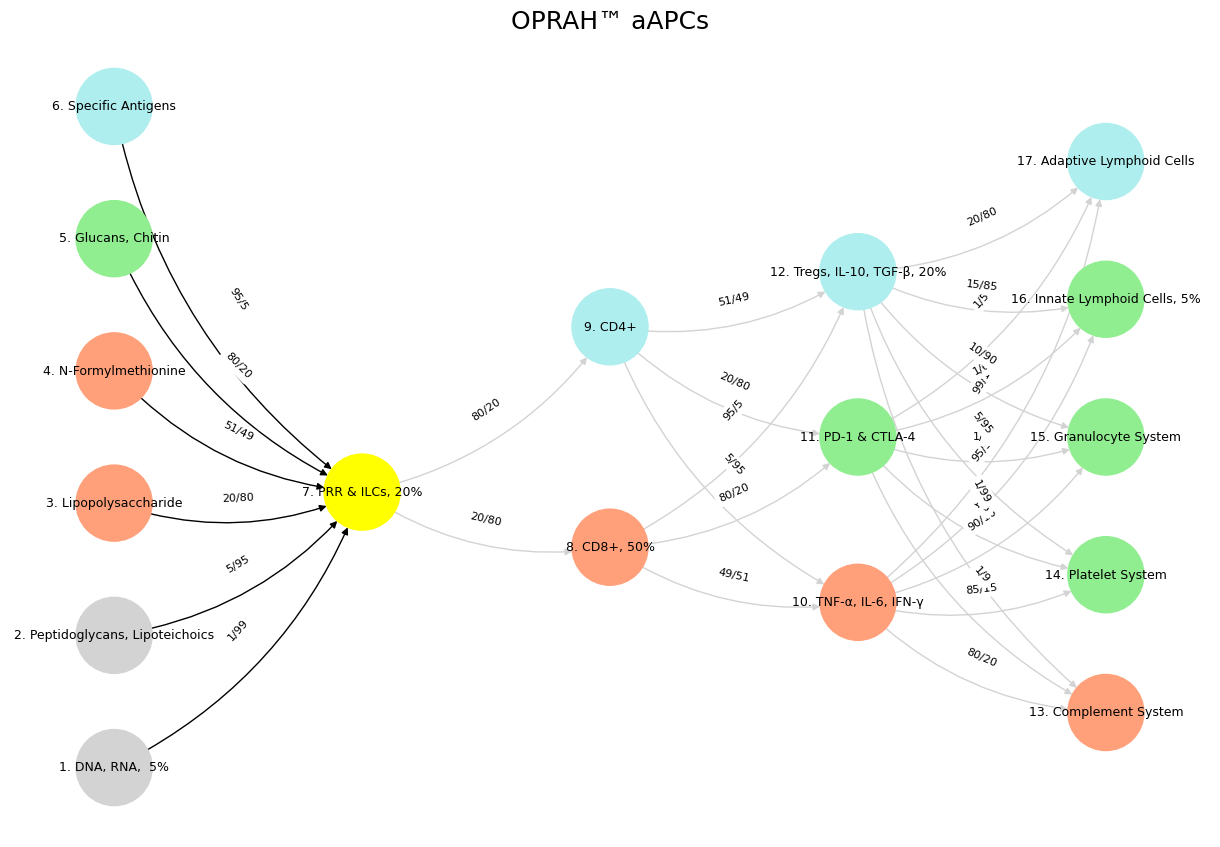
Fig. 37 While neural biology inspired neural networks in machine learning, the realization that scaling laws apply so beautifully to machine learning has led to a divergence in the process of generation of intelligence. Biology is constrained by the Red Queen, whereas mankind is quite open to destroying the Ecosystem-Cost function for the sake of generating the most powerful AI.#

