Dancing in Chains#
Below is an essay making a case for mismatch repair, weaving in some of the provided context and drawing parallels to biological systems, while avoiding bullet points and adhering to a paragraph-only structure as requested. The essay reframes the Murdoch family dynamics as a metaphor for cellular repair mechanisms and builds a scientific argument for mismatch repair’s importance, incorporating the given formula w = 1/[1 + X/Y] where relevant.
The Murdoch family retreat of 2010, as recounted by James Murdoch, offers a compelling lens through which to view the concept of mismatch repair—not in the psychological sense, though the parallels are striking, but in the biological one. Rupert Murdoch, the patriarch, gathered his children in an attempt to address festering familial tensions, hiring a therapist to mend what he perceived as misalignments in their relationships. Yet, as James described, the effort backfired, leaving everyone more estranged than before—a “car crash” of miscommunication and competing agendas. In cellular biology, mismatch repair serves a similar purpose: to correct errors that arise during DNA replication, ensuring the integrity of the genetic code. When replication machinery mispairs nucleotides—say, an adenine with a cytosine instead of a thymine—the mismatch repair system steps in to detect and rectify the mistake. Without it, the consequences ripple outward, much like the unresolved rivalries in the Murdoch clan, threatening the stability of the organism as mutations accumulate over time.

Fig. 32 Freedom in Fetters—a Princely Freedom. Chopin, the last of the modern musicians, who gazed at and worshipped beauty, like Leopardi; Chopin, the Pole, the inimitable (none that came before or after him has a right to this name)—Chopin had the same princely punctilio in convention (grammar, space) that Raphael shows in the use of the simplest traditional colours. The only difference is that Chopin applies them not to colour but to melodic and rhythmic traditions (prosody, time). He admitted the validity of these traditions because he was born under the sway of etiquette. But in these fetters he plays and dances as the freest and daintiest of spirits, and, be it observed, he does not spurn the chain. Source: Human All-Too-Human Part II#
The importance of mismatch repair becomes even clearer when we consider the signal-to-noise ratio, a concept encapsulated in the formula w = 1/[1 + X/Y], where X/Y represents the noise-to-signal ratio or, in immunological terms, the variant-to-conserved epitope balance. In the Murdoch family, the “noise” of personal agendas—James’s skepticism, Rupert’s control, Liz’s business interests—overwhelmed the “signal” of familial unity the retreat aimed to reinforce. In DNA replication, noise manifests as errors introduced by polymerase slippage or environmental damage, while the signal is the faithful transmission of the original sequence. Mismatch repair proteins, such as MutS and MutL in bacteria or their eukaryotic homologs MSH and MLH, act as mediators, binding to distorted DNA helices where base pairs don’t align. They recruit excision machinery to remove the erroneous segment and resynthesize it correctly. The weighting factor “w” in the formula reflects the system’s efficiency: as noise (X) increases relative to signal (Y), the effectiveness of repair diminishes, underscoring the need for a robust mechanism to keep X/Y low. Without mismatch repair, cells risk losing the signal entirely, much as the Murdochs lost any hope of cohesion when their mediation failed.
This repair system’s role extends beyond mere error correction to the broader context of cellular survival and adaptation, a stakes-driven drama not unlike the succession struggles at News Corp. In the retreat, James noted his father’s tendency to “manipulate them against each other,” a tactic that mirrors how unrepaired DNA mismatches can destabilize a genome, pitting cellular processes against themselves. Uncorrected mismatches during replication can lead to strand breaks or stalled replication forks, triggering apoptosis if damage exceeds repair capacity. More insidiously, persistent errors fuel mutagenesis, increasing the likelihood of cancer—colorectal cancer, for instance, is strongly linked to defects in mismatch repair genes like MSH2 and MLH1. The Murdoch children’s retreat, ending in alienation, reflects a failure to reconcile competing elements; in biology, a failure of mismatch repair similarly leaves a cell vulnerable, unable to reconcile its genetic identity with the errors threatening it. The stakes are high: data suggest that mismatch repair defects account for up to 15-20% of sporadic colorectal cancers, a testament to its protective role.
Full of sound and fury,
Signifying nothing
– Macbeth
Moreover, mismatch repair’s efficiency ties into the immune system’s ability to distinguish self from nonself, a dynamic hinted at in the neural network code’s layered architecture—from pathogen recognition (Suis) to effector responses (Deviens). The retreat’s therapist, outmatched by the family’s complexities, parallels the overwhelm of an immune system facing unchecked genomic instability. When mismatch repair falters, microsatellite instability arises, producing novel peptide sequences—variant epitopes—that the immune system may flag as foreign. Here, the X/Y ratio shifts: conserved epitopes (signal) diminish as variant ones (noise) proliferate, taxing immune surveillance. CD4+ and CD8+ T-cells, as modeled in the network, must then contend with this imbalance, potentially leading to chronic inflammation or immune evasion by tumors. The Murdoch family’s inability to align their narratives mirrors a cell’s struggle to maintain its identity amid rising noise, reinforcing why mismatch repair is indispensable for genomic and immunological harmony.
In advocating for mismatch repair, we see a system that, like a skilled mediator, preserves order amid chaos—whether in a cell or a family dynasty. The Murdoch retreat’s collapse illustrates what happens when repair fails: entropy prevails, and unity erodes. Biologically, mismatch repair ensures that DNA replication’s fidelity keeps pace with life’s demands, maintaining a low X/Y ratio to safeguard the signal of heredity. Its absence invites catastrophe—cancer, immune dysfunction, cellular collapse—much as the Murdoch children’s unresolved conflicts invited ongoing strife. As James reflected, some things are better left unpicked; yet in genetics, picking at mismatches is precisely what sustains life. The case for mismatch repair is thus unassailable: it is the unsung guardian of stability, quietly correcting errors so the system, cellular or familial, can endure.
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['DNA, RNA, 5%', 'Peptidoglycans, Lipoteichoics', 'Lipopolysaccharide', 'N-Formylmethionine', "Glucans, Chitin", 'Specific Antigens'],
'Voir': ['PRR & ILCs, 20%'],
'Choisis': ['CD8+, 50%', 'CD4+'],
'Deviens': ['TNF-α, IL-6, IFN-γ', 'PD-1 & CTLA-4', 'Tregs, IL-10, TGF-β, 20%'],
"M'èléve": ['Complement System', 'Platelet System', 'Granulocyte System', 'Innate Lymphoid Cells, 5%', 'Adaptive Lymphoid Cells']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['PRR & ILCs, 20%'],
'paleturquoise': ['Specific Antigens', 'CD4+', 'Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'],
'lightgreen': ["Glucans, Chitin", 'PD-1 & CTLA-4', 'Platelet System', 'Innate Lymphoid Cells, 5%', 'Granulocyte System'],
'lightsalmon': ['Lipopolysaccharide', 'N-Formylmethionine', 'CD8+, 50%', 'TNF-α, IL-6, IFN-γ', 'Complement System'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights
def define_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
('PRR & ILCs, 20%', 'CD8+, 50%'): '20/80',
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD8+, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
('CD8+, 50%', 'PD-1 & CTLA-4'): '80/20',
('CD8+, 50%', 'Tregs, IL-10, TGF-β, 20%'): '95/5',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD4+', 'PD-1 & CTLA-4'): '20/80',
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Platelet System'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Granulocyte System'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Innate Lymphoid Cells, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Adaptive Lymphoid Cells'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Platelet System'): '1/8',
('PD-1 & CTLA-4', 'Granulocyte System'): '1/7',
('PD-1 & CTLA-4', 'Innate Lymphoid Cells, 5%'): '1/6',
('PD-1 & CTLA-4', 'Adaptive Lymphoid Cells'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Platelet System'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Granulocyte System'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Define edges to be highlighted in black
def define_black_edges():
return {
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD4+', 'PD-1 & CTLA-4'): '20/80',
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
}
# Calculate node positions
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
black_edges = define_black_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
edge_colors = []
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
edge_colors.append('black' if (source, target) in black_edges else 'lightgrey')
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color=edge_colors,
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™: Nonself & the Salient Network", fontsize=18)
plt.show()
# Run the visualization
visualize_nn()
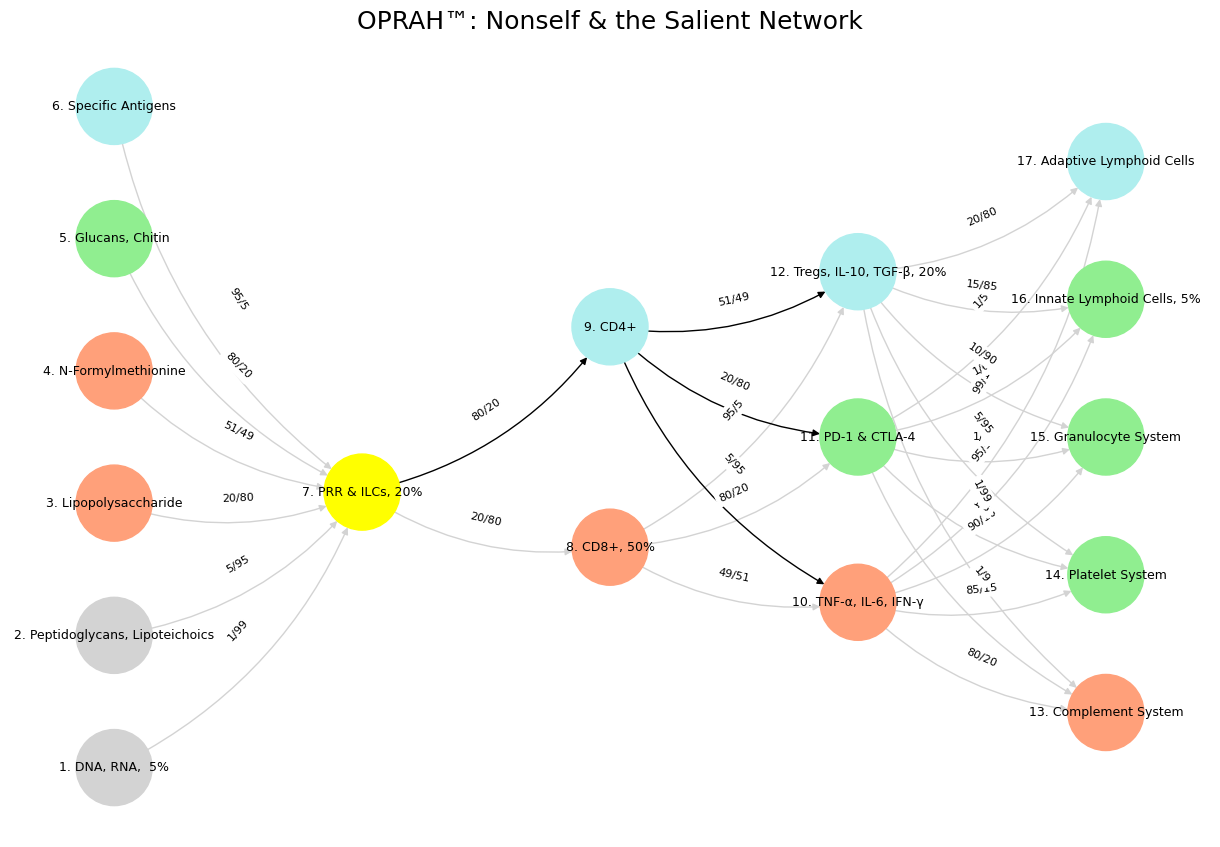
Fig. 33 G1-G3: Ganglia & N1-N5 Nuclei. These are cranial nerve, dorsal-root (G1 & G2); basal ganglia, thalamus, hypothalamus (N1, N2, N3); and brain stem and cerebelum (N4 & N5).#

