Apollo & Dionysus#
🪙 🎲 🎰 🐜 🗡️ 🪖 🛡️#
The immune system’s metaphors of “self,” “nonself,” and “preservation” offer a compelling lens through which to view the essence of truce negotiations in adversarial relationships, whether they be between nations like Ukraine and Russia or Hamas and Israel, or within familial dynasties like the Murdochs’ “Project Harmony.” In the immune system, self is defined by MHC-I, a marker of original identity rooted in the parental DNA, ensuring that the body recognizes its own cells as safe. Nonself, conversely, is anything foreign—detected by T-cell receptors (TCR) or antigen-presenting cells (APCs)—that triggers a defensive response unless modulated by mechanisms like PD-L1, which negotiates a truce to prevent overreaction. Preservation, then, is the system’s ultimate goal: maintaining the integrity of the self against threats while adapting to changing conditions. Applied to tokenized transactional relationships on a blockchain-like ledger—where identity, trust, and value are distributed and immutable—this framework suggests that negotiations hinge on defining what constitutes the “self” worth preserving and how to coexist with the “nonself” without mutual destruction.
🎭#

Fig. 19 Pattern Recognition Receptors: Difference in Language and Communication. Republicans have honed a way to viscerally connect with white heterosexual protestant English-speaking men. It’s the intelligence of a tribe whose pedigree has encoded the trial and errors of its ancestors into a subtext that is accessible. Democrats fail to see their opponents’ text as mere bait and commit a related grammatical error of overstating the power of grammar (think DEI, 21st century feminist, millenial, and Gen-Z vocabulary of outrage.. a flourishing school of resentment). The immune system, like the neural and endocrine systems, have high-risk, high-error heuristics-bias sentinal signaling#
🌊 🏄🏾#
In the context of Ukraine-Russia or Hamas-Israel, the “self” might be each party’s national or cultural identity, encoded in historical narratives and territorial claims, much like DNA in a cell. The “nonself” is the adversary, perceived as a threat to survival, triggering an immune-like escalation—war—unless a truce can tokenize a transactional relationship. Here, blockchain metaphors shine: a truce could be a smart contract, a verifiable ledger of commitments (e.g., ceasefire terms, hostage releases, or aid flows) that both sides agree to uphold, preserving their core “self” while acknowledging the other’s existence. The essence of these negotiations lies in mismatch repair—correcting misalignments in expectations or violations (like Israel’s aid blockades or Hamas’s refusal to extend phase one of a ceasefire)—to avoid total breakdown. Yet, as with the immune system, this preservation is temporary, tied to the current “parental” generation of leaders or conditions. Once they pass—say, Putin or Netanyahu—the offspring (new leaders, new regimes) inherit only about 50% of the prior DNA, reshaping the body politic and its priorities, rendering the old truce obsolete.
🤺 💵 🦘#
The Murdoch family saga, particularly “Project Harmony,” mirrors this dynamic with a twist of legacy. Rupert, at 93, embodies the parental DNA of News Corp, his MHC-I defining the conservative “self” he’s fought to preserve against the “nonself” of his more liberal-leaning children, like James. “Project Harmony” could be seen as his attempt to tokenize a truce— a blockchain of succession where shares and influence are distributed (albeit unevenly, with a 1:3 split favoring his vision)—to maintain his legacy’s integrity. Yet, like an immune response, this is a negotiation fraught with mismatch madness: his offspring, only partially aligned with his DNA, are poised to redefine the company’s identity once he’s gone. The existential angst here echoes King Lear—Rupert, dividing his kingdom, risks losing it to a new generation’s vision, a liberal shift he can scarcely fathom. Why does he bother? Because, as with the immune system, preservation of the self is instinctual while the parent lives; legacy is the body politic’s survival mechanism, even if it’s doomed to evolve beyond his control.
🏇 🧘🏾♀️ 🪺 🎶 🛌#
Extending this to warring nations, the metaphor reveals a brutal truth: truces are not about permanent peace but about preserving the current “self” against the “nonself” for as long as the parental template—be it a leader, ideology, or power structure—endures. Ukraine and Russia negotiate not for eternal harmony but to tokenize a transactional pause, preserving their respective DNAs (sovereignty, influence) amid mismatch repair (e.g., territorial disputes). Hamas and Israel do the same, with each ceasefire phase a ledger entry to delay annihilation. Legacy drives this—Putin’s Russia, Netanyahu’s Israel, or Rupert’s empire all cling to their founding codes. But the immune system teaches us that once the parent dies, the offspring’s 50% divergence births a new reference point. For nations, this might mean new leadership or generational shifts; for the Murdochs, it’s a post-Rupert News Corp. In all cases, the angst of legacy is futile yet inevitable—nature guarantees change, and no truce, no matter how tokenized, can outlast the death of its DNA. King Lear’s lesson is universal: the old guard fights for control, but the new world waits, indifferent.
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['DNA, RNA, 5%', 'Peptidoglycans, Lipoteichoics', 'Lipopolysaccharide', 'N-Formylmethionine', "Glucans, Chitin", 'Specific Antigens'],
'Voir': ['PRR & ILCs, 20%'],
'Choisis': ['CD8+, 50%', 'CD4+'],
'Deviens': ['TNF-α, IL-6, IFN-γ', 'PD-1 & CTLA-4', 'Tregs, IL-10, TGF-β, 20%'],
"M'èléve": ['Complement System', 'Platelet System', 'Granulocyte System', 'Innate Lymphoid Cells, 5%', 'Adaptive Lymphoid Cells']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['PRR & ILCs, 20%'],
'paleturquoise': ['Specific Antigens', 'CD4+', 'Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'],
'lightgreen': ["Glucans, Chitin", 'PD-1 & CTLA-4', 'Platelet System', 'Innate Lymphoid Cells, 5%', 'Granulocyte System'],
'lightsalmon': ['Lipopolysaccharide', 'N-Formylmethionine', 'CD8+, 50%', 'TNF-α, IL-6, IFN-γ', 'Complement System'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights
def define_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
('PRR & ILCs, 20%', 'CD8+, 50%'): '20/80',
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD8+, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
('CD8+, 50%', 'PD-1 & CTLA-4'): '80/20',
('CD8+, 50%', 'Tregs, IL-10, TGF-β, 20%'): '95/5',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD4+', 'PD-1 & CTLA-4'): '20/80',
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Platelet System'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Granulocyte System'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Innate Lymphoid Cells, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Adaptive Lymphoid Cells'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Platelet System'): '1/8',
('PD-1 & CTLA-4', 'Granulocyte System'): '1/7',
('PD-1 & CTLA-4', 'Innate Lymphoid Cells, 5%'): '1/6',
('PD-1 & CTLA-4', 'Adaptive Lymphoid Cells'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Platelet System'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Granulocyte System'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Define edges to be highlighted in black
def define_black_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
}
# Calculate node positions
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
black_edges = define_black_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
edge_colors = []
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
edge_colors.append('black' if (source, target) in black_edges else 'lightgrey')
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color=edge_colors,
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™ & ARNTC", fontsize=18)
plt.show()
# Run the visualization
visualize_nn()
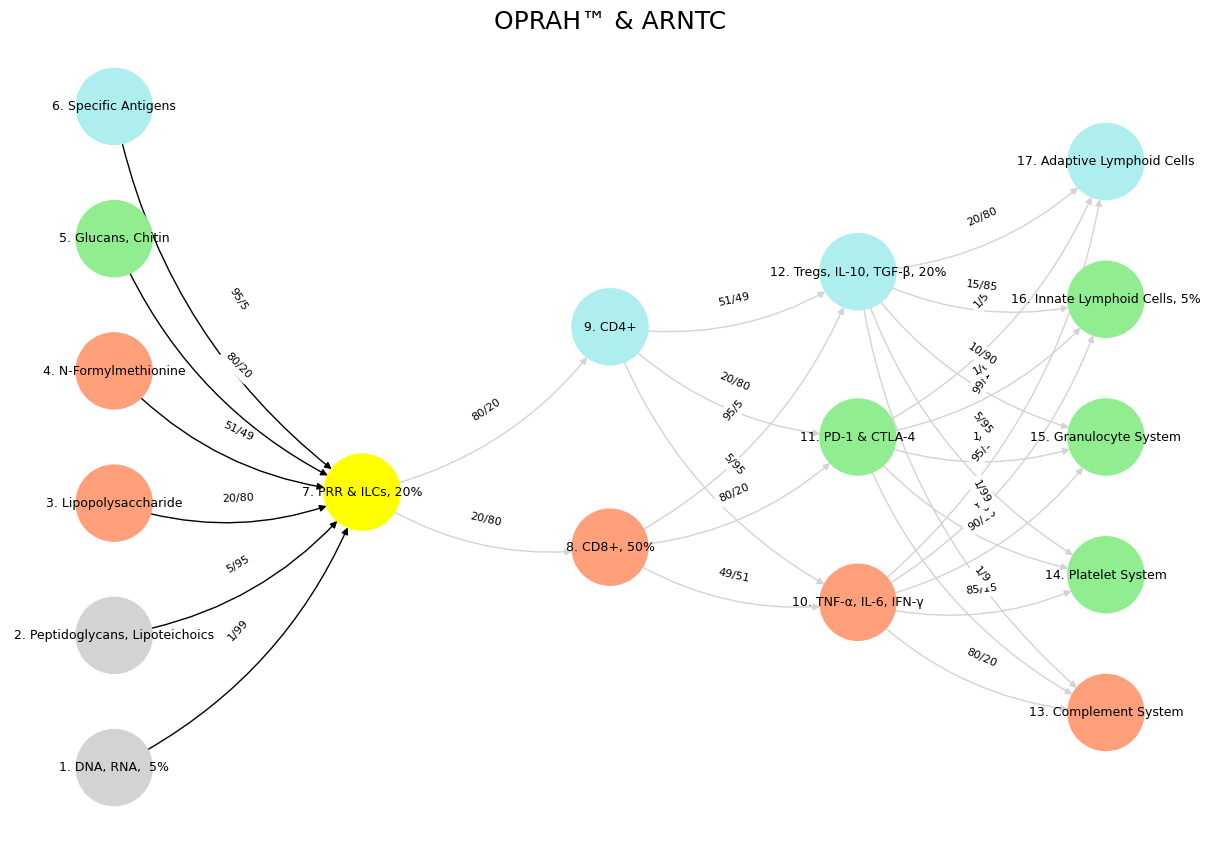
Fig. 20 Nonself. This is anything foreign—Epitope.MHC-I or MHC-II complex, detected by T-cell receptors (TCR) or antigen-presenting cells (APCs)—that triggers a defensive response unless modulated by mechanisms like PD-L1, which negotiates a truce to prevent overreaction.#

