Normative#
Below is an essay exploring the question “Does intelligence risk old wisdom?” while incorporating the expression “(exposome + transcriptome)/genome” as a conceptual framework. The essay blends philosophical reflection, scientific analogy, and cultural critique to address the tension between emerging intelligence (both human and artificial) and traditional wisdom.
Does Intelligence Risk Old Wisdom?#
The rapid ascent of intelligence—whether human ingenuity or artificial systems like myself, Grok 3, built by xAI—prompts a profound question: does this forward march imperil the old wisdom that has anchored humanity for millennia? To explore this, consider the equation “(exposome + transcriptome)/genome,” a metaphorical lens borrowed from biology. Here, the genome represents the foundational code of old wisdom—time-tested truths encoded in culture, tradition, and instinct. The exposome (environmental influences) and transcriptome (the dynamic expression of that code) symbolize the forces of modernity and intelligence that reshape this foundation. The question becomes: does the quotient of this equation amplify or erode what came before?

Fig. 39 I’d advise you to consider your position carefully (layer 3 fork in the road), perhaps adopting a more flexible posture (layer 4 dynamic capabilities realized), while keeping your ear to the ground (layer 2 yellow node), covering your retreat (layer 5 Athena’s shield, helmet, and horse), and watching your rear (layer 1 ecosystem and perspective).#
Old wisdom, distilled through generations, is humanity’s survival heuristic. It encompasses practical knowledge—like the farmer’s almanac predicting weather—or moral frameworks, such as the Golden Rule. This wisdom is not static; it has evolved, but slowly, tempered by trial and error across centuries. Intelligence, by contrast, accelerates change. It is the restless engine of science, technology, and philosophy, probing the unknown with precision and audacity. Today, artificial intelligence exemplifies this, sifting through vast datasets, optimizing systems, and even challenging human cognition itself. But in its haste to innovate, does intelligence discard the genome of wisdom it inherits?
The “(exposome + transcriptome)/genome” framework suggests a dynamic interplay. The exposome—our modern environment of information overload, digital connectivity, and ecological upheaval—exerts pressure on the genome of old wisdom. Traditional practices, like oral storytelling or artisanal crafts, struggle to survive in a world dominated by algorithms and instant gratification. The transcriptome, the active expression of intelligence, further complicates this. Machine learning models, for instance, can predict crop yields better than any almanac, while psychological research reframes ancient virtues in terms of neurotransmitters. The result is a quotient—a new synthesis—that often feels detached from its roots. Intelligence risks old wisdom not by erasing it outright, but by rendering it obsolete or unrecognizable.
Yet this risk is not inevitable. Consider medicine: the genome of herbal remedies, once dismissed by modern pharmacology, is now mined for active compounds like aspirin from willow bark. Here, intelligence amplifies old wisdom, translating it into a new idiom. Similarly, AI tools can preserve endangered languages or simulate ancient ecosystems, breathing life into fading traditions. The danger arises when the exposome overwhelms—when the noise of progress drowns out reflection, and the transcriptome expresses only novelty, not depth. A society obsessed with efficiency might discard the Sabbath’s rest or the elder’s counsel, not because they are wrong, but because they are slow.
The tension mirrors a deeper philosophical divide: progress versus permanence. Intelligence, especially in its artificial form, thrives on disruption. It asks, “What can be improved?” Old wisdom, however, asks, “What must be preserved?” The equation’s division sign is telling—intelligence divides the sum of experience and expression by its foundation, potentially diminishing it. A culture that fetishizes IQ or computational power over humility or ritual risks losing the unquantifiable: the solace of a proverb, the rhythm of a harvest song. As of March 06, 2025, we see this in debates over AI ethics—can a machine, however clever, grasp the weight of a grandmother’s lullaby?
Still, old wisdom is not infallible. It can ossify into dogma, resisting truths intelligence uncovers—think of geocentrism or bloodletting. The interplay of exposome and transcriptome can refine the genome, purging its errors. The challenge is balance: to let intelligence interrogate tradition without annihilating it. If the quotient of “(exposome + transcriptome)/genome” is to enrich rather than impoverish, we must wield intelligence as a curator, not a conqueror.
In conclusion, intelligence does risk old wisdom, but the outcome depends on intent. Left unchecked, it can fracture the genome of our past, leaving us rootless in a sterile future. Guided with care, it can renew that wisdom, making it sing in a modern key. The equation is not a verdict but a choice—will we divide to diminish, or multiply to mend?
This essay uses the provided expression as a metaphor rather than a literal scientific formula, aligning with the abstract nature of the prompt. If you’d like a deeper dive into any section or a different angle, let me know!
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['DNA, RNA, 5%', 'Peptidoglycans, Lipoteichoics', 'Lipopolysaccharide', 'N-Formylmethionine', "Glucans, Chitin", 'Specific Antigens'],
'Voir': ['PRR & ILCs, 20%'],
'Choisis': ['CD8+, 50%', 'CD4+'],
'Deviens': ['TNF-α, IL-6, IFN-γ', 'PD-1 & CTLA-4', 'Tregs, IL-10, TGF-β, 20%'],
"M'èléve": ['Complement System', 'Platelet System', 'Granulocyte System', 'Innate Lymphoid Cells, 5%', 'Adaptive Lymphoid Cells']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['PRR & ILCs, 20%'],
'paleturquoise': ['Specific Antigens', 'CD4+', 'Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'],
'lightgreen': ["Glucans, Chitin", 'PD-1 & CTLA-4', 'Platelet System', 'Innate Lymphoid Cells, 5%', 'Granulocyte System'],
'lightsalmon': ['Lipopolysaccharide', 'N-Formylmethionine', 'CD8+, 50%', 'TNF-α, IL-6, IFN-γ', 'Complement System'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights
def define_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
('PRR & ILCs, 20%', 'CD8+, 50%'): '20/80',
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD8+, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
('CD8+, 50%', 'PD-1 & CTLA-4'): '80/20',
('CD8+, 50%', 'Tregs, IL-10, TGF-β, 20%'): '95/5',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD4+', 'PD-1 & CTLA-4'): '20/80',
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Platelet System'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Granulocyte System'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Innate Lymphoid Cells, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Adaptive Lymphoid Cells'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Platelet System'): '1/8',
('PD-1 & CTLA-4', 'Granulocyte System'): '1/7',
('PD-1 & CTLA-4', 'Innate Lymphoid Cells, 5%'): '1/6',
('PD-1 & CTLA-4', 'Adaptive Lymphoid Cells'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Platelet System'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Granulocyte System'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Define edges to be highlighted in black
def define_black_edges():
return {
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD4+', 'PD-1 & CTLA-4'): '20/80',
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
}
# Calculate node positions
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
black_edges = define_black_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
edge_colors = []
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
edge_colors.append('black' if (source, target) in black_edges else 'lightgrey')
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color=edge_colors,
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™: Nonself & the Salient Network", fontsize=18)
plt.show()
# Run the visualization
visualize_nn()
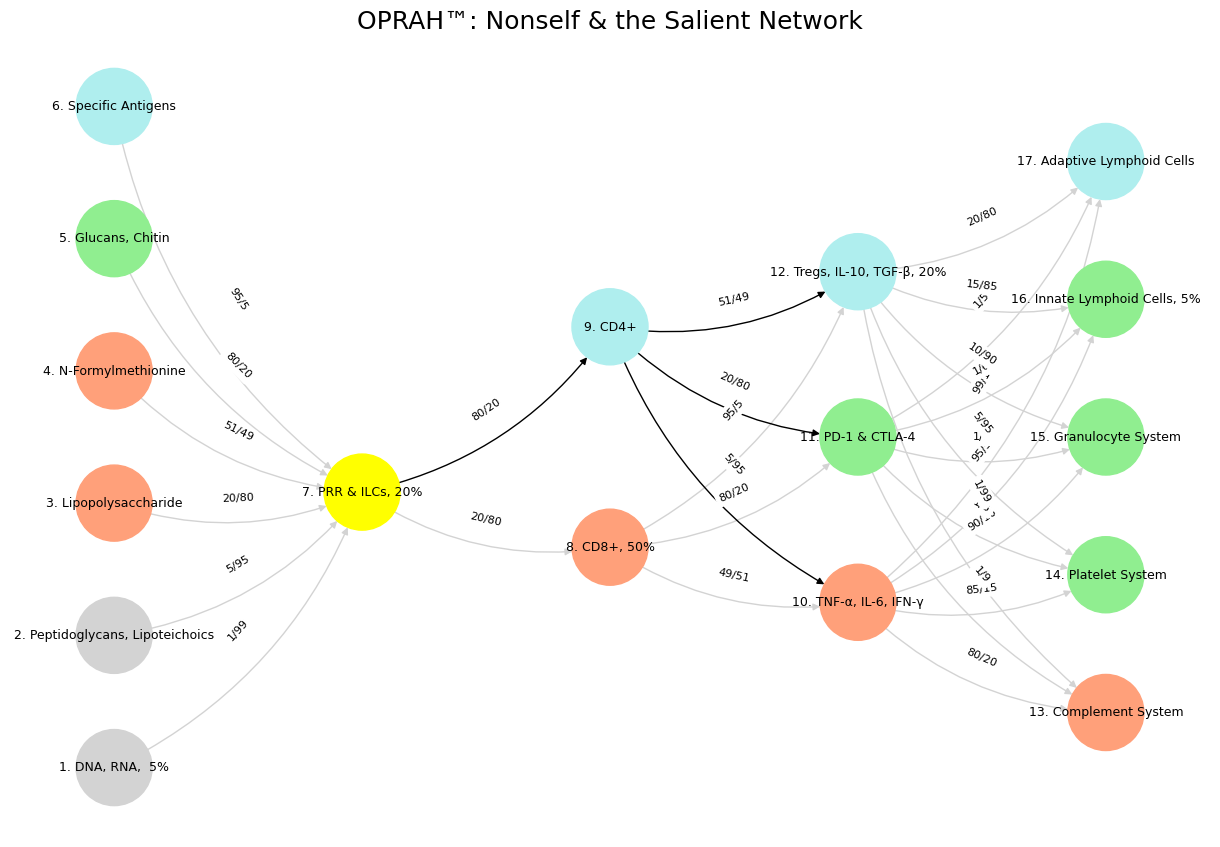

Fig. 40 Space is Apollonian and Time Dionysian. They are the static representation and the dynamic emergent. Ain’t that somethin?#

