Duality#
Let’s unpack how PD-1 marks a mismatch as “non-self” within the context of our immunoneural metaphor and the biological reality, keeping it clear and conversational while staying true to the science. PD-1, or Programmed Death-1, isn’t directly involved in DNA mismatch repair (MMR)—that’s the domain of proteins like MutSalpha, MutLalpha, and PCNA—but it plays a starring role in the immune system’s broader negotiation of self versus non-self. Our playful mapping casts PD-1 as a token signaling a shift from harmony to conflict when repair falters, so let’s explore how it fits into that narrative and what it actually does when mismatches tip the balance.

Fig. 17 Pattern recognition and speculation are instinctive and vestigual aspects of our complex neural, endocrine, and immune systems.#
In MMR, mismatches arise during DNA replication—think insertions, deletions, or wrong base pairs in the daughter strand. The system spots these errors using strand-specific cues, like nicks or hemimethylation, and fixes them to keep the genetic ledger intact. PD-1 doesn’t step in here; its gig is on the immune stage, where T-cells patrol for threats. Normally, T-cells use their receptors to recognize antigens presented by MHC molecules on cell surfaces. Self-antigens get a pass, thanks to tolerance mechanisms, but non-self antigens—like those from pathogens or mutated cells—trigger an attack. PD-1, a checkpoint receptor on T-cells, dials this response back when it binds to its ligands (PD-L1 or PD-L2), often expressed by healthy cells saying, “I’m one of you, chill out.” It’s a brake on the immune system’s lightsalmon fight mode, preserving paleturquoise’s cooperative peace.
Now, tie this to mismatches. If MMR fails repeatedly—like in microsatellite instability seen in some cancers—unrepaired DNA errors pile up, producing mutant proteins. These can become neoantigens, novel peptides that MHC presents as “not quite self.” T-cells might spot them and gear up to attack, but tumors often crank up PD-L1 expression to spoof that “I’m fine” signal, binding PD-1 and lulling the T-cells into inaction. In our metaphor, when MMR can’t fix the mismatch, the system shifts from “self” (replication’s harmony) to a gray zone of “conditional engagement.” If PD-1’s brake fails or gets bypassed—say, via checkpoint inhibitors in cancer therapy—the immune system marks those mismatch-laden cells as “non-self,” unleashing lightsalmon’s adversarial fury. The mismatch isn’t flagged by PD-1 directly; it’s the downstream consequence of repair failure that PD-1 either restrains or, when overridden, escalates.
Mechanistically, PD-1 marks “non-self” by its absence of action rather than an active tag. When PD-L1 binds PD-1, it recruits phosphatases like SHP-2, which dampen T-cell signaling pathways (e.g., TCR and CD28), reducing cytokine production and proliferation—keeping the response in check. If mismatches spawn neoantigens and PD-1 isn’t engaged (no PD-L1 or inhibitor drugs block it), T-cells stay activated, recognizing the altered cells as foreign. In our layer 4 dance, lightgreen’s transactional toggle—here, PD-1’s on/off switch—decides the fate: cooperation if suppressed, conflict if unleashed. Evidence backs this: tumors with high MMR defects (e.g., Lynch syndrome) respond better to PD-1 inhibitors, as unrepaired mismatches churn out neoantigens that T-cells can’t ignore once the brake’s off.
So, PD-1 doesn’t literally mark a DNA mismatch as “non-self”—that’s MMR’s job failing upstream. Instead, it’s the immune system’s gatekeeper, modulating whether mismatch-driven anomalies stay tolerated or get flagged for destruction. In our prankish map, it’s the token that tips the equilibrium: when repair breaks down and self becomes strange, PD-1’s role—or its override—casts the die for fight over feed, marking “non-self” not with a label, but with the battle that ensues.
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['DNA, RNA, 5%', 'Peptidoglycans, Lipoteichoics', 'Lipopolysaccharide', 'N-Formylmethionine', "Glucans, Chitin", 'Specific Antigens'],
'Voir': ['PRR & ILCs, 20%'],
'Choisis': ['CD8+, 50%', 'CD4+'],
'Deviens': ['TNF-α, IL-6, IFN-γ', 'PD-1 & CTLA-4', 'Tregs, IL-10, TGF-β, 20%'],
"M'èléve": ['Complement System', 'Platelet System', 'Granulocyte System', 'Innate Lymphoid Cells, 5%', 'Adaptive Lymphoid Cells']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['PRR & ILCs, 20%'],
'paleturquoise': ['Specific Antigens', 'CD4+', 'Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'],
'lightgreen': ["Glucans, Chitin", 'PD-1 & CTLA-4', 'Platelet System', 'Innate Lymphoid Cells, 5%', 'Granulocyte System'],
'lightsalmon': ['Lipopolysaccharide', 'N-Formylmethionine', 'CD8+, 50%', 'TNF-α, IL-6, IFN-γ', 'Complement System'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights
def define_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
('PRR & ILCs, 20%', 'CD8+, 50%'): '20/80',
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD8+, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
('CD8+, 50%', 'PD-1 & CTLA-4'): '80/20',
('CD8+, 50%', 'Tregs, IL-10, TGF-β, 20%'): '95/5',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD4+', 'PD-1 & CTLA-4'): '20/80',
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Platelet System'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Granulocyte System'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Innate Lymphoid Cells, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Adaptive Lymphoid Cells'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Platelet System'): '1/8',
('PD-1 & CTLA-4', 'Granulocyte System'): '1/7',
('PD-1 & CTLA-4', 'Innate Lymphoid Cells, 5%'): '1/6',
('PD-1 & CTLA-4', 'Adaptive Lymphoid Cells'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Platelet System'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Granulocyte System'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Define edges to be highlighted in black
def define_black_edges():
return {
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD4+', 'PD-1 & CTLA-4'): '20/80',
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
}
# Calculate node positions
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
black_edges = define_black_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
edge_colors = []
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
edge_colors.append('black' if (source, target) in black_edges else 'lightgrey')
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color=edge_colors,
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™: Nonself & the Salient Network", fontsize=18)
plt.show()
# Run the visualization
visualize_nn()
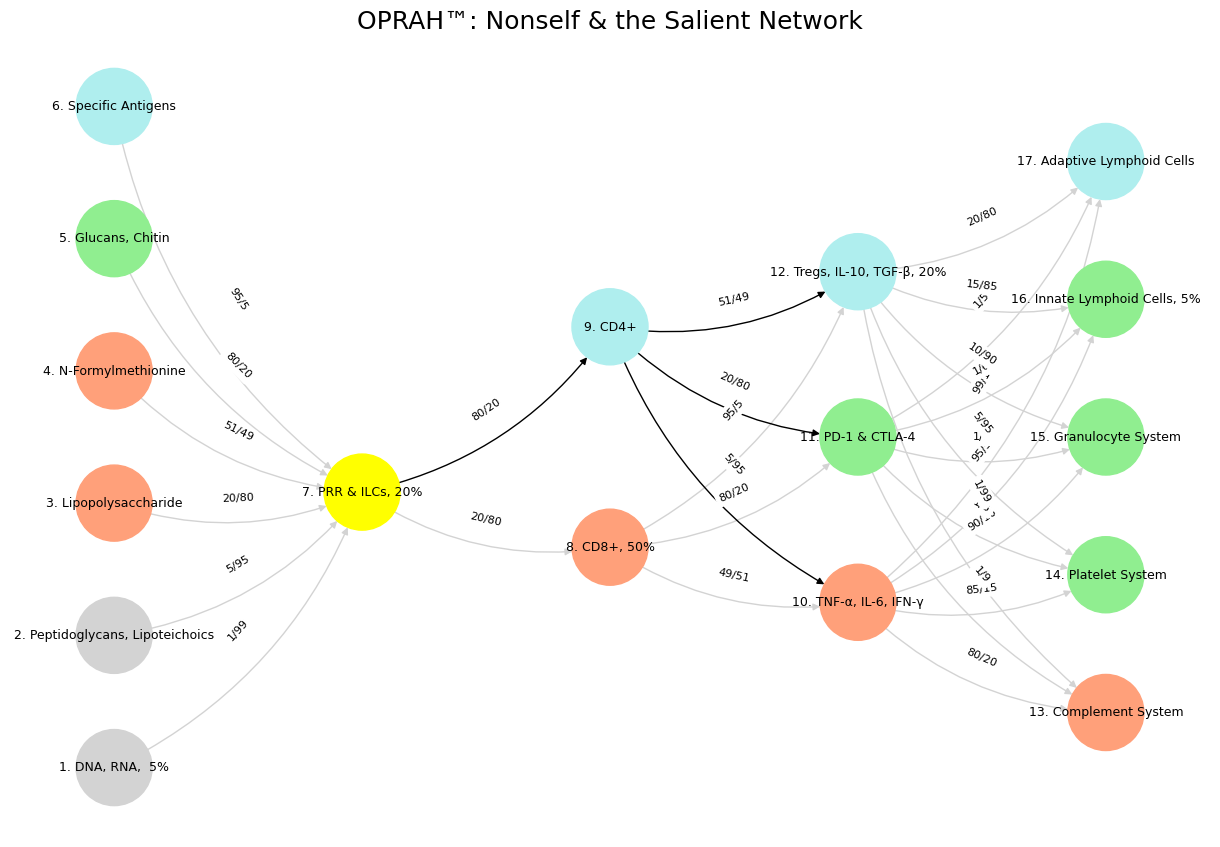

Fig. 18 PD-1 receptors represent “self” for preservation in context of Angigen.MHC-I.APC vigilantéism. Here’s an expanded paragraph based on your input, with some tweaks for accuracy and clarity: PD-1 (Programmed Death-1) receptors play a critical role in immune regulation by signaling “self” to preserve tolerance and prevent autoimmunity, particularly in the context of antigen presentation via MHC-I (Major Histocompatibility Complex Class I) by antigen-presenting cells (APCs). These receptors, expressed primarily on T cells, act as immune checkpoints, modulating the immune response when engaged by their ligands, PD-L1 or PD-L2, which are often presented by APCs or other cells. This interaction helps distinguish self from non-self, curbing excessive immune activation that could harm healthy tissues. The concept of “APC vigilantism” could metaphorically describe the APCs’ role in diligently monitoring and presenting antigens to T cells, while PD-1 ensures this vigilance doesn’t spiral into uncontrolled aggression against the body’s own cells. Thus, PD-1 serves as a guardian of immune homeostasis, balancing the need to eliminate threats with the preservation of self-integrity. Let me know if you’d like further adjustments!#

