Ecosystem#
Let’s dive deeper into our whimsical yet oddly illuminating mapping of the immune system onto the nervous system, shifting our gaze to layer 4, where the interplay of dynamic equilibria takes center stage. Here, lightsalmon embodies the sympathetic nervous system’s fight, flight, and fright responses—those adrenal surges that scream adversarial engagement. Paleturquoise, in contrast, channels the parasympathetic’s gentle hum of feed, breed, and sleep, a cooperative vibe that soothes and sustains. Then there’s lightgreen, the presynaptic autonomic ganglion, toggling and transacting like a neural switchboard, mediating between the two with a transactional flair. These colors aren’t just aesthetic choices; they’re our playful proxies for the push and pull of biological systems, reflecting how DNA’s ancestral whispers ripple through replication and repair to shape life’s ongoing negotiation with itself and its environment.

Fig. 10 Hubris. The function of hubris in tragedy is to interrogate static vs. dynamic equilibria. Should we expect Mozartean cadences or eternally recurrent Wagnarian melodies? Should the trial and error ever end? Should we ever decelerate –rest– from the Red Queen Hypothesis? Do immutable laws from the cosmos demand immutable laws in our physics, biology, and intelligence if ever our loss is to be optimized?#
After DNA replication—think of it as the “breed” phase where the self doubles down on itself—mismatch repair steps in like a meticulous editor, ensuring fidelity in the script. This system, known as MMR, is a molecular proofreader, spotting and fixing erroneous insertions, deletions, and mis-incorporations that sneak in during replication or recombination. It’s strand-specific, a critical detail: the newly minted daughter strand, prone to errors, needs to be distinguished from the trusty parental template. In gram-negative bacteria, transient hemimethylation does the trick—the parental strand’s methylation tags it as the original, while the daughter lags behind, unmarked. In eukaryotes, though, the mechanism’s murkier. It’s thought that nicks in the newly synthesized lagging strand—gaps left before DNA ligase seals them—act as signposts, guiding MMR machinery to the right target. Recent evidence even suggests these nicks pepper the leading strand too, adding a twist to the tale.
Zooming in, the transactional dance of mismatch repair gets even more intricate. Nicks aren’t just flaws; they’re docking stations. Research shows they’re where replication factor C (RFC) loads the replication sliding clamp, PCNA, in a precise, orientation-specific way—one face of this donut-shaped protein cozying up to the 3’-OH end of the nick. Once loaded, PCNA becomes the conductor, directing the MutLalpha endonuclease to snip the daughter strand when a mismatch is flagged by MutSalpha or MutSbeta. It’s a handshake of molecular precision, a transactional imperative ensuring the system keeps dealing fairly. PD-1 enters the scene here as a token of continued negotiation—a checkpoint that, when MMR falters, shifts the dynamic from “self” (replication’s harmony) to “conditional engagement” (repair’s triage) and, if unchecked, to “non-self” (an adversarial clash).
Now, let’s weave this into our layer 4 framework. The sympathetic lightsalmon flares when mismatch repair fails, and PD-1 signals “fight”—an immune response kicking into gear against perceived threats, a breakdown of self-recognition spiraling into conflict. Paleturquoise’s parasympathetic calm aligns with successful repair, a return to “breed” and “feed,” where the system cooperates with itself to maintain harmony. Lightgreen’s presynaptic ganglion, ever the transactional mediator, toggles between these states, balancing the adversarial and cooperative through PCNA’s careful choreography. It’s a dynamic equilibrium, a teeter-totter of molecular and neural metaphors where DNA’s fidelity mirrors the nervous system’s own balancing act—fight or rest, clash or connect, all mediated by the quiet transactions that keep the whole show running.
This playful mapping isn’t just a thought experiment; it’s a lens on how systems, biological or otherwise, negotiate their edges. The immune system’s MMR and the nervous system’s autonomic toggles share a logic of recognition and response, of distinguishing self from other and adapting accordingly. Lightsalmon, paleturquoise, and lightgreen become more than colors—they’re the hues of life’s perpetual bargaining, where every nick, every repair, every signal is a move in the game of equilibrium. And in that game, PD-1’s role as a token reminds us: even in failure, there’s a chance to deal anew, to shift the balance, and to keep the dance alive.
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['DNA, RNA, 5%', 'Peptidoglycans, Lipoteichoics', 'Lipopolysaccharide', 'N-Formylmethionine', "Glucans, Chitin", 'Specific Antigens'],
'Voir': ['PRR & ILCs, 20%'],
'Choisis': ['CD8+, 50%', 'CD4+'],
'Deviens': ['TNF-α, IL-6, IFN-γ', 'PD-1 & CTLA-4', 'Tregs, IL-10, TGF-β, 20%'],
"M'èléve": ['Complement System', 'Platelet System', 'Granulocyte System', 'Innate Lymphoid Cells, 5%', 'Adaptive Lymphoid Cells']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['PRR & ILCs, 20%'],
'paleturquoise': ['Specific Antigens', 'CD4+', 'Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'],
'lightgreen': ["Glucans, Chitin", 'PD-1 & CTLA-4', 'Platelet System', 'Innate Lymphoid Cells, 5%', 'Granulocyte System'],
'lightsalmon': ['Lipopolysaccharide', 'N-Formylmethionine', 'CD8+, 50%', 'TNF-α, IL-6, IFN-γ', 'Complement System'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights
def define_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
('PRR & ILCs, 20%', 'CD8+, 50%'): '20/80',
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD8+, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
('CD8+, 50%', 'PD-1 & CTLA-4'): '80/20',
('CD8+, 50%', 'Tregs, IL-10, TGF-β, 20%'): '95/5',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD4+', 'PD-1 & CTLA-4'): '20/80',
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Platelet System'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Granulocyte System'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Innate Lymphoid Cells, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Adaptive Lymphoid Cells'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Platelet System'): '1/8',
('PD-1 & CTLA-4', 'Granulocyte System'): '1/7',
('PD-1 & CTLA-4', 'Innate Lymphoid Cells, 5%'): '1/6',
('PD-1 & CTLA-4', 'Adaptive Lymphoid Cells'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Platelet System'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Granulocyte System'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Define edges to be highlighted in black
def define_black_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
}
# Calculate node positions
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
black_edges = define_black_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
edge_colors = []
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
edge_colors.append('black' if (source, target) in black_edges else 'lightgrey')
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color=edge_colors,
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™: Task-Positive Network", fontsize=18)
plt.show()
# Run the visualization
visualize_nn()
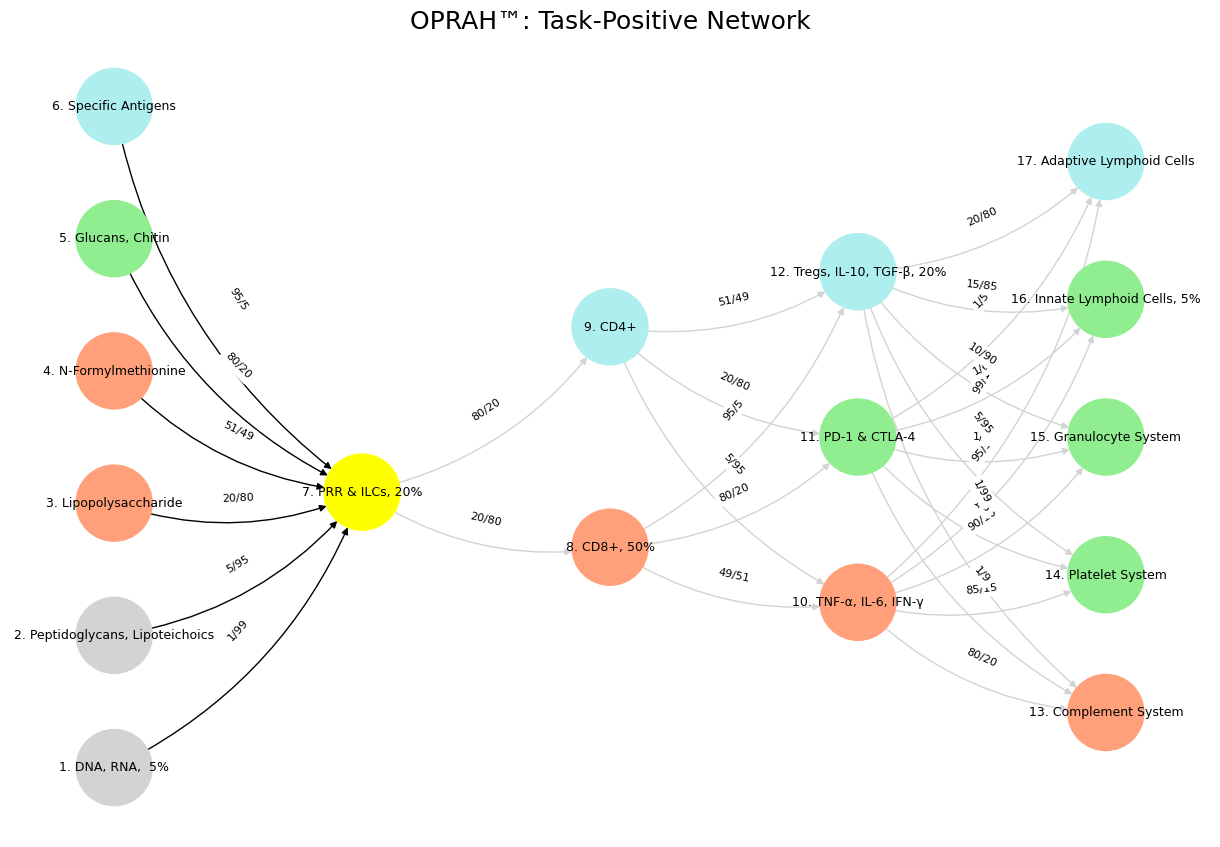

Fig. 11 For the eyes of the Lord run to and fro throughout the whole earth, to shew himself strong in the behalf of them whose heart is perfect toward him. Herein thou hast done foolishly: therefore from henceforth thou shalt have wars. Source: 2 Chronicles 16: 8-9. The grammar of these visuals is plain: there’s a space & time for the cooperative rhythm, transactional, and adversarial.#

