Traditional#
🪙 🎲 🎰 🔍 👁️ 🐍#
Your clarification steers the essay toward a more specific interpretation of “dancing in chains,” tying Macbeth’s wisdom explicitly to the noise/signal paradigm and the equation (exposome + transcriptome)/genome = fury/significance. Here, Rupert Murdoch’s actions during the 2010 family retreat, as recounted by James, become a lens to explore whether he misses the insight that excessive noise—environmental and expressive pressures—overwhelms significance, mirroring Macbeth’s descent into futile chaos. “Dancing in chains” shifts from Nietzsche’s artistic constraint to a biological and Shakespearean metaphor: the struggle to maintain meaning (signal or significance) amid disruption (noise or fury), as expressed in the exposome (external influences), transcriptome (cellular responses), and genome (core identity). This essay, in paragraphs only, investigates if Rupert’s failure to manage this ratio reflects a complete oversight of Macbeth’s wisdom.

Fig. 37 I’d advise you to consider your position carefully (layer 3 fork in the road), perhaps adopting a more flexible posture (layer 4 dynamic capabilities realized), while keeping your ear to the ground (layer 2 yellow node), covering your retreat (layer 5 Athena’s shield, helmet, and horse), and watching your rear (layer 1 ecosystem and perspective).#
👂🏾#
Rupert Murdoch’s 2010 retreat emerges as a crucible where noise drowned signal, a dynamic James Murdoch’s “car crash” description lays bare. In your framing, noise/signal equates to (exposome + transcriptome)/genome, where the exposome—external stressors like Rupert’s domineering presence and the therapist’s intervention—plus the transcriptome—the family’s reactive squabbles and emotional outbursts—overwhelmed the genome, their shared lineage and purpose. Macbeth’s wisdom, distilled in “full of sound and fury, signifying nothing,” warns that unchecked noise (fury) erodes significance (meaning); Rupert, by forcing reconciliation, amplified the exposome’s chaos and triggered a transcriptome of resentment, diluting the signal of familial unity. His intent to repair mismatches—rivalries threatening the dynasty—mirrors a cell’s effort to correct DNA errors, but his approach piled fury atop fury, suggesting he missed the lesson that noise, left unmastered, renders the dance of power hollow.
🌊 🏄🏾#
This noise/signal imbalance finds a stark parallel in biology, where the exposome (environmental mutagens) and transcriptome (gene expression shifts) challenge the genome’s integrity. Mismatch repair keeps this ratio in check, excising errors to preserve the signal—much as a wise leader might quiet discord to sustain purpose. In Macbeth, the protagonist’s ambition becomes his exposome, his paranoid decrees the transcriptome, together burying the genome of his rightful rule; the result is noise overwhelming signal, a life of fury with no lasting significance. Rupert’s retreat, with its manipulative undertones, parallels this: James noted his father “manipulated them against each other,” a fury of control that drowned the significance of their shared legacy. The formula w = 1/[1 + X/Y] quantifies this failure—as X (noise: exposome + transcriptome) surged relative to Y (signal: genome), w approached zero, marking Rupert’s repair attempt as ineffective, blind to Macbeth’s cautionary tale.
🗡️ 🪖 🛡️#
Yet Rupert’s dance within chains hints at partial awareness, complicating the claim he “completely misses” this wisdom. Macbeth operates within fate’s fetters, his fury a misstep within constraint; Rupert, too, worked within the dynasty’s structure, his exposome of influence and transcriptome of directives aimed at preserving the genome of his empire. “Dancing in chains” here suggests a constrained struggle to balance noise and signal—Rupert sensed the mismatches but misjudged their repair, letting fury dominate. Biologically, a cell overburdened by exposome and transcriptome noise risks mutation when repair falters; socially, Rupert’s family risked fracture, the retreat’s fallout a microsatellite instability of trust. He grasped the chains—the power dynamics—but not the dance, the finesse to keep (exposome + transcriptome)/genome low, aligning with Macbeth’s insight only in its tragic outcome, not its foresight.
🏇 🧘🏾♀️ 🔱 🎶 🛌#
The stakes of this oversight loom large, both in Shakespeare and genetics, illuminating Rupert’s shortfall. Macbeth’s kingdom collapses as fury overtakes significance, his signal lost to noise; a genome swamped by exposome and transcriptome noise faces cancer or collapse, as in mismatch repair-deficient tumors. The Murdoch retreat didn’t end the dynasty, but its signal—familial cohesion—faded amid the fury James recounted, a significance diminished by noise. Rupert’s failure to see this ratio’s tipping point—where (exposome + transcriptome)/genome = fury/significance—marks his disconnect from Macbeth’s wisdom. He danced, but without grace, amplifying noise over signal until the retreat signified little beyond strife. Not a complete miss, perhaps, but a stumble so profound that Macbeth’s lesson—noise unchecked is meaning undone—remained tragically out of reach.
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['DNA, RNA, 5%', 'Peptidoglycans, Lipoteichoics', 'Lipopolysaccharide', 'N-Formylmethionine', "Glucans, Chitin", 'Specific Antigens'],
'Voir': ['PRR & ILCs, 20%'],
'Choisis': ['CD8+, 50%', 'CD4+'],
'Deviens': ['TNF-α, IL-6, IFN-γ', 'PD-1 & CTLA-4', 'Tregs, IL-10, TGF-β, 20%'],
"M'èléve": ['Complement System', 'Platelet System', 'Granulocyte System', 'Innate Lymphoid Cells, 5%', 'Adaptive Lymphoid Cells']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['PRR & ILCs, 20%'],
'paleturquoise': ['Specific Antigens', 'CD4+', 'Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'],
'lightgreen': ["Glucans, Chitin", 'PD-1 & CTLA-4', 'Platelet System', 'Innate Lymphoid Cells, 5%', 'Granulocyte System'],
'lightsalmon': ['Lipopolysaccharide', 'N-Formylmethionine', 'CD8+, 50%', 'TNF-α, IL-6, IFN-γ', 'Complement System'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights
def define_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
('PRR & ILCs, 20%', 'CD8+, 50%'): '20/80',
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD8+, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
('CD8+, 50%', 'PD-1 & CTLA-4'): '80/20',
('CD8+, 50%', 'Tregs, IL-10, TGF-β, 20%'): '95/5',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD4+', 'PD-1 & CTLA-4'): '20/80',
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Platelet System'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Granulocyte System'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Innate Lymphoid Cells, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Adaptive Lymphoid Cells'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Platelet System'): '1/8',
('PD-1 & CTLA-4', 'Granulocyte System'): '1/7',
('PD-1 & CTLA-4', 'Innate Lymphoid Cells, 5%'): '1/6',
('PD-1 & CTLA-4', 'Adaptive Lymphoid Cells'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Platelet System'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Granulocyte System'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Define edges to be highlighted in black
def define_black_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
}
# Calculate node positions
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
black_edges = define_black_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
edge_colors = []
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
edge_colors.append('black' if (source, target) in black_edges else 'lightgrey')
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color=edge_colors,
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™ aAPCs", fontsize=18)
plt.show()
# Run the visualization
visualize_nn()
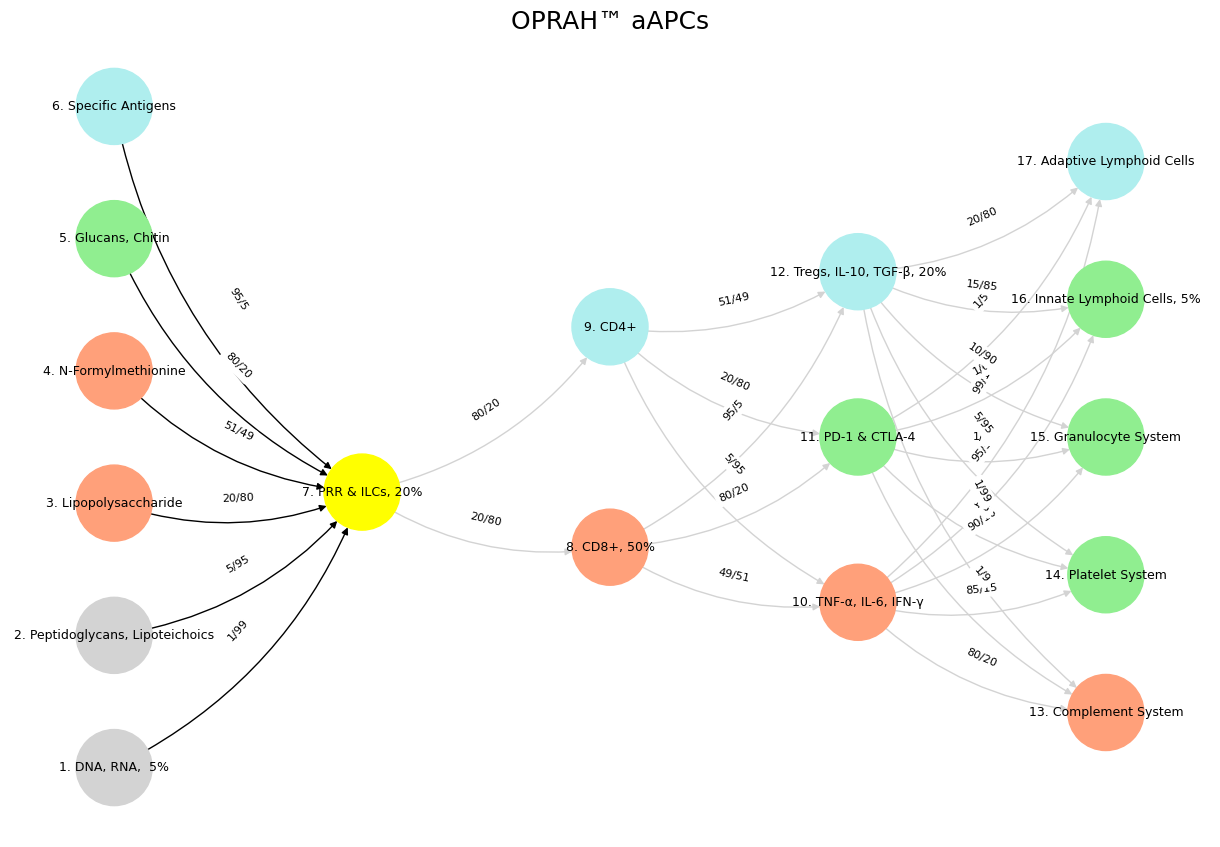

Fig. 38 While neural biology inspired neural networks in machine learning, the realization that scaling laws apply so beautifully to machine learning has led to a divergence in the process of generation of intelligence. Biology is constrained by the Red Queen, whereas mankind is quite open to destroying the Ecosystem-Cost function for the sake of generating the most powerful AI.#

