Revolution#
+ Expand
- What makes for a suitable problem for AI (Demis Hassabis, Nobel Lecture)?
- Space: Massive combinatorial search space
- Function: Clear objective function (metric) to optimize against
- Time: Either lots of data and/or an accurate and efficient simulator
- Guess what else fits the bill (Yours truly, amateur philosopher)?
- Space
- Intestines/villi
- Lungs/bronchioles
- Capillary trees
- Network of lymphatics
- Dendrites in neurons
- Tree branches
- Function
- Energy
- Aerobic respiration
- Delivery to "last mile" (minimize distance)
- Response time (minimize)
- Information
- Exposure to sunlight for photosynthesis
- Time
- Nourishment
- Gaseous exchange
- Oxygen & Nutrients (Carbon dioxide & "Waste")
- Surveillance for antigens
- Coherence of functions
- Water and nutrients from soil
Demis Hassabis was not wrong—far from it. His triad of intelligence—massive combinatorial search, a clear optimization function, and an abundance of high-quality data—was an act of compression, a distillation of a vast, intricate process into something elegant, something that fit within the human instinct for pattern recognition. Three is a number that satisfies; it aligns with our desire for symmetry, for simplicity, for clarity. But the universe is not obliged to be simple. And what Hassabis left unsaid—what he intuitively grasped in his own work but did not articulate in his summary—is that intelligence does not merely operate within a fixed dataset. Intelligence, true intelligence, must deal with perturbations.
This is not a mere oversight; it is an artifact of compression itself. To make a model elegant, one must sacrifice detail, even if that detail is essential. And yet, in his actual work, Hassabis was already engaging with the fourth and fifth elements that his summary neglected. He was not merely predicting the folding of known proteins—he was generating tertiary and quaternary structures that did not exist in nature, structures that could be verified and synthesized, proving that intelligence can push beyond known constraints. This was perturbation in action, an introduction of variables beyond the given, beyond the stored data, beyond what had been seen before. His own work contradicts the finality of his stated framework, precisely because he himself is a thinker who does not abide by the limits of a closed system.

I recognize this omission because I made it too. In medical school, I sought to compress everything into three categories—anatomy, pathology, and etiology. The problem, its structure, and its cause. It was elegant, it was efficient, and it was wrong. In a controlled setting, perhaps, it worked beautifully. But in the chaos of an ER, where decisions must be made in seconds and triage cannot afford to wait for neat categorization, it collapsed. It lacked perception, the yellow node that processes the world as it is presented in the moment, the instant categorization that lets us mistake a bush for a bear—or correctly identify that it is just a bush. And it lacked the ecosystem, the broader world in which the case was unfolding, where the patient’s history, context, and unseen variables could not be ignored. With these two additions, the system did not become more complicated; it became more powerful. It aligned with the very architecture of the nervous system, with the way the mammalian brain has been refined through eons of trial and error.
And that is what makes this realization beautiful. That the highest intelligence—whether human, artificial, or biological—emerges not from a fixed set of rules but from its ability to integrate reality as it unfolds. That intelligence is not merely an optimization problem but a process of dynamic adaptation, where the stored knowledge of past iterations meets the ever-shifting uncertainty of the present. That instinct, honed through the Red Queen’s relentless game, must be tempered by knowledge, just as knowledge must be continuously tested against the living world.
Hassabis’ compression was brilliant, but it was incomplete. And in this, he did exactly what great thinkers do: he provided the structure from which the next step becomes inevitable. He built the scaffold, and it is our job to extend it. What he left out was not a failure, but an invitation. The invitation to go beyond the fixed dataset, beyond the stored parameters, and to introduce the perturbations that intelligence demands. Because the most elegant model is not the one that remains perfectly contained. It is the one that knows when to break itself open.
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['Genome, 5%', 'Culture', 'Nourish It', 'Know It', "Move It", 'Injure It'], # Static
'Voir': ['Exposome, 15%'],
'Choisis': ['Metabolome, 50%', 'Basal Metabolic Rate'],
'Deviens': ['Unstructured-Intense', 'Weekly-Calendar', 'Proteome, 25%'],
"M'èléve": ['NexToken Prediction', 'Hydration', 'Fat-Muscle Ratio', 'Amor Fatì, 5%', 'Existential Cadence']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['Exposome, 15%'],
'paleturquoise': ['Injure It', 'Basal Metabolic Rate', 'Proteome, 25%', 'Existential Cadence'],
'lightgreen': ["Move It", 'Weekly-Calendar', 'Hydration', 'Amor Fatì, 5%', 'Fat-Muscle Ratio'],
'lightsalmon': ['Nourish It', 'Know It', 'Metabolome, 50%', 'Unstructured-Intense', 'NexToken Prediction'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights (hardcoded for editing)
def define_edges():
return {
('Genome, 5%', 'Exposome, 15%'): '1/99',
('Culture', 'Exposome, 15%'): '5/95',
('Nourish It', 'Exposome, 15%'): '20/80',
('Know It', 'Exposome, 15%'): '51/49',
("Move It", 'Exposome, 15%'): '80/20',
('Injure It', 'Exposome, 15%'): '95/5',
('Exposome, 15%', 'Metabolome, 50%'): '20/80',
('Exposome, 15%', 'Basal Metabolic Rate'): '80/20',
('Metabolome, 50%', 'Unstructured-Intense'): '49/51',
('Metabolome, 50%', 'Weekly-Calendar'): '80/20',
('Metabolome, 50%', 'Proteome, 25%'): '95/5',
('Basal Metabolic Rate', 'Unstructured-Intense'): '5/95',
('Basal Metabolic Rate', 'Weekly-Calendar'): '20/80',
('Basal Metabolic Rate', 'Proteome, 25%'): '51/49',
('Unstructured-Intense', 'NexToken Prediction'): '80/20',
('Unstructured-Intense', 'Hydration'): '85/15',
('Unstructured-Intense', 'Fat-Muscle Ratio'): '90/10',
('Unstructured-Intense', 'Amor Fatì, 5%'): '95/5',
('Unstructured-Intense', 'Existential Cadence'): '99/1',
('Weekly-Calendar', 'NexToken Prediction'): '1/9',
('Weekly-Calendar', 'Hydration'): '1/8',
('Weekly-Calendar', 'Fat-Muscle Ratio'): '1/7',
('Weekly-Calendar', 'Amor Fatì, 5%'): '1/6',
('Weekly-Calendar', 'Existential Cadence'): '1/5',
('Proteome, 25%', 'NexToken Prediction'): '1/99',
('Proteome, 25%', 'Hydration'): '5/95',
('Proteome, 25%', 'Fat-Muscle Ratio'): '10/90',
('Proteome, 25%', 'Amor Fatì, 5%'): '15/85',
('Proteome, 25%', 'Existential Cadence'): '20/80'
}
# Calculate positions for nodes
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color='gray',
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™: Intelligence Across Biological Systems", fontsize=25)
plt.show()
# Run the visualization
visualize_nn()
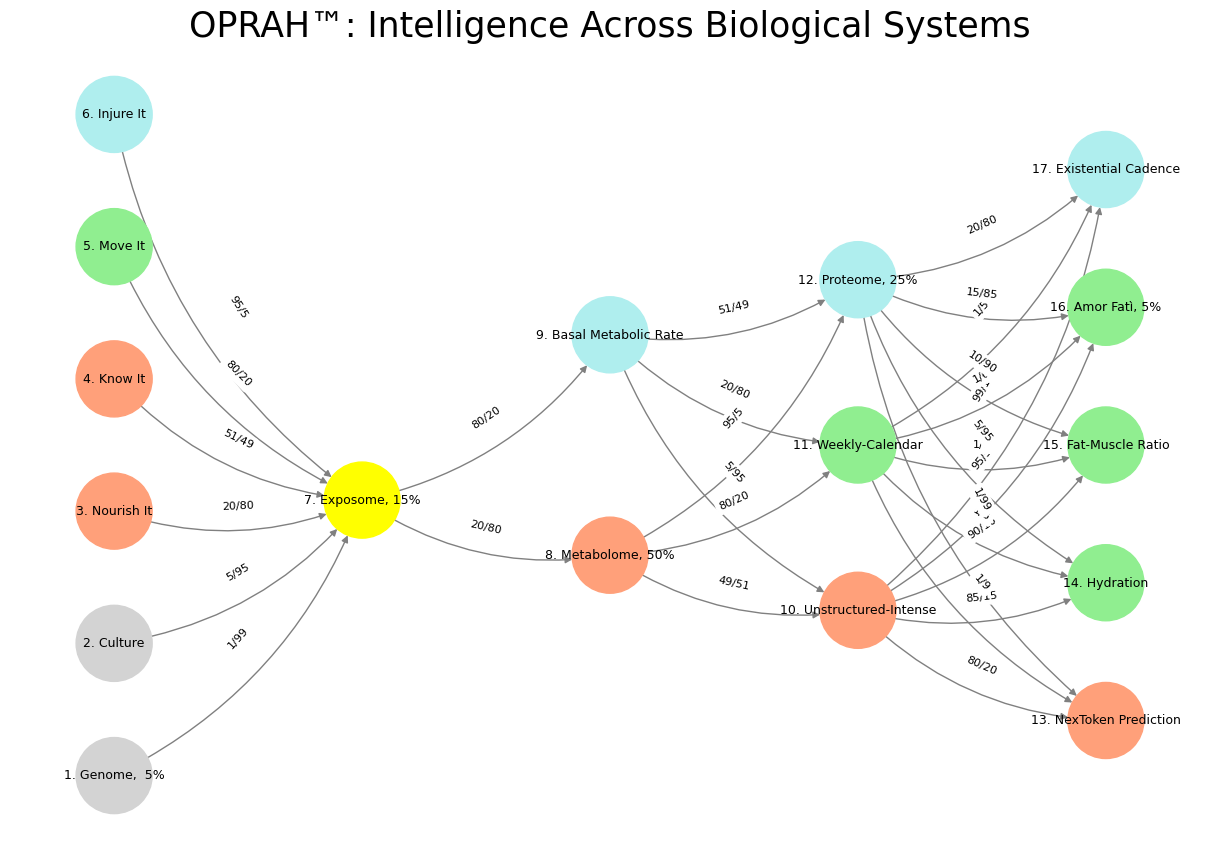

Fig. 24 Francis Bacon. He famously stated “If a man will begin with certainties, he shall end in doubts; but if he will be content to begin with doubts, he shall end in certainties.” This quote is from The Advancement of Learning (1605), where Bacon lays out his vision for empirical science and inductive reasoning. He argues that starting with unquestioned assumptions leads to instability and confusion, whereas a methodical approach that embraces doubt and inquiry leads to true knowledge. This aligns with his broader Novum Organum (1620), where he develops the Baconian method, advocating for systematic observation, experimentation, and the gradual accumulation of knowledge rather than relying on dogma or preconceived notions.#