Transvaluation#
The OPRAH™ system’s five-network framework—Suis, Voir, Choisis, Deviens, and M’élève—presents itself as a bold synthesis of immunology and neuroanatomy, promising a unified model where the body’s defenses and the mind’s operations mirror each other in a seamless, layered hierarchy. It’s an ambitious claim: that these systems aren’t just analogous but fundamentally intertwined, sharing a computational and cognitive logic across their five stages. Yet this framework, while intricate, leans heavily on speculative leaps and overextended parallels, glossing over gaps in evidence and biological specificity. The result is a model that dazzles with symmetry but strains under scrutiny, raising questions about its coherence and practical utility.

Fig. 21 Digital Twin Avatar. It can be used to scale ones personhood, give lectures in more than one place, at more than one time, in more than one langugage. The agentic AIs are the translators and perhaps the stage is set for physical MESH AIs to embody these in the future.#
Suis, the first layer, ties the pericentral network’s reflexes to the innate immune system’s pattern recognition receptors, framing both as immediate, pre-conscious responses. The idea is compelling—pain withdrawal and pathogen detection as two sides of the same survival coin, driven by speed over deliberation. But the comparison stumbles in its generality. Neural reflexes are localized, millisecond-fast circuits, while innate immunity, though rapid, unfolds over minutes to hours, involving complex cellular cascades. The framework’s insistence on their equivalence glosses over these temporal and mechanistic disparities, presenting a tidy alignment that feels more poetic than precise. It assumes a shared “evolutionary programming” without grappling with how these systems diverged in their actual machinery, risking a conflation of metaphor with fact.
Voir, the next step, links the dorsal network’s goal-directed motion to inflammatory signaling, casting both as tactical escalations from raw reaction to purposeful action. The parallel is seductive—cytokines rallying immune cells like the brain steering a hand toward a target. Yet inflammation is a blunt instrument, often spilling beyond its intent, while dorsal stream actions are finely tuned, spatially precise. The framework paints this as a smooth handoff, but the analogy buckles under the weight of their differences. Inflammation’s systemic chaos contrasts sharply with the dorsal network’s focused control, and claiming they serve the same “directed” purpose stretches the model thin. It’s a leap that prioritizes narrative flow over the messy reality of biological processes, sidelining the distinct constraints each system faces.
Choisis, aligning the lateral network’s working memory with immune memory, posits a shared capacity for learning and adaptation—antigen recognition mirroring cognitive recall. This is the framework’s strongest pitch: both systems refine themselves through experience, compressing the past into future readiness. But the analogy falters in scale and substance. Immune memory is a distributed, cellular archive, etched in lymphocyte populations over years, while working memory is fleeting, neuronally encoded, and context-specific. The model’s assertion of “true intelligence” here feels grandiose, equating a slow, probabilistic immune process with the brain’s rapid, flexible cognition. It’s an overreach that blurs critical distinctions, banking on a vague notion of “refinement” to bridge a gap that biology doesn’t fully support.

Fig. 22 Salient Network: Midcingulo-Insular Network. The framework’s real limit is its refusal to admit how shallow these parallels run—immune and neural systems don’t “converge” beyond basic problem-solving, and dressing them in tech jargon doesn’t deepen the link. OPRAH™ maps a compelling comparison, but its sharpness cuts only skin-deep, leaving the messy guts of biology untouched.#
Deviens, pairing the medial network’s self-awareness with immune self-regulation, aims to balance tolerance and aggression—PD-1 and Tregs echoing introspection and emotional control. The concept of a “self” negotiated across both domains is intriguing, but the execution falters. Immune tolerance is a molecular checkpoint system, not a conscious deliberation, while medial network functions like social cognition involve layered, subjective experiences far beyond immune mechanics. The framework’s talk of “harmony or chaos” overdramatizes a regulatory process into a psychological drama, projecting a unity that doesn’t hold. Autoimmunity and mental dysregulation may share superficial parallels as failures of balance, but their causes and expressions are so divergent that the comparison risks being more rhetorical than revealing.
M’élève, the top layer, merges the midcingulo-insular salience network with immune homeostasis, framing both as arbiters of priority—deciding what warrants a response. The salience network’s emotional filtering and the complement system’s threat assessment do share a selective logic, but the framework pushes this too far. The brain’s salience is a nuanced, moment-to-moment judgment, while immune homeostasis involves slower, systemic adjustments—platelets and granulocytes don’t “feel” threats the way the insula weighs significance. Calling this a unified “great arbiter” inflates a functional overlap into a philosophical stance, ignoring how these systems prioritize at vastly different scales. The model’s elegance here comes at the cost of precision, favoring a grand narrative over grounded analysis.
The OPRAH™ framework’s core flaw lies in its relentless pursuit of symmetry. It assumes neural and immune systems don’t just collaborate but operate as a single, mirrored intelligence, a feedback loop so tight that distinctions dissolve. This is a captivating vision, but it sacrifices specificity for sweep, papering over the gritty details—time scales, mechanisms, evolutionary pressures—that define these systems’ real interactions. Its predictive power, touted as a breakthrough, remains untested, resting on assertions of pattern recognition that lack concrete examples. The rejection of “outdated three-layer models” feels more like a rhetorical flourish than a substantiated advance, leaving the five-layer structure as a hypothesis dressed up as a revelation. It’s a model that sings with ambition but struggles to harmonize with the discordant notes of biological reality.
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['DNA, RNA, 5%', 'Peptidoglycans, Lipoteichoics', 'Lipopolysaccharide', 'N-Formylmethionine', "Glucans, Chitin", 'Specific Antigens'],
'Voir': ['PRR & ILCs, 20%'],
'Choisis': ['CD8+, 50%', 'CD4+'],
'Deviens': ['TNF-α, IL-6, IFN-γ', 'PD-1 & CTLA-4', 'Tregs, IL-10, TGF-β, 20%'],
"M'èléve": ['Complement System', 'Platelet System', 'Granulocyte System', 'Innate Lymphoid Cells, 5%', 'Adaptive Lymphoid Cells']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['PRR & ILCs, 20%'],
'paleturquoise': ['Specific Antigens', 'CD4+', 'Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'],
'lightgreen': ["Glucans, Chitin", 'PD-1 & CTLA-4', 'Platelet System', 'Innate Lymphoid Cells, 5%', 'Granulocyte System'],
'lightsalmon': ['Lipopolysaccharide', 'N-Formylmethionine', 'CD8+, 50%', 'TNF-α, IL-6, IFN-γ', 'Complement System'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights
def define_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
('PRR & ILCs, 20%', 'CD8+, 50%'): '20/80',
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD8+, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
('CD8+, 50%', 'PD-1 & CTLA-4'): '80/20',
('CD8+, 50%', 'Tregs, IL-10, TGF-β, 20%'): '95/5',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD4+', 'PD-1 & CTLA-4'): '20/80',
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Platelet System'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Granulocyte System'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Innate Lymphoid Cells, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Adaptive Lymphoid Cells'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Platelet System'): '1/8',
('PD-1 & CTLA-4', 'Granulocyte System'): '1/7',
('PD-1 & CTLA-4', 'Innate Lymphoid Cells, 5%'): '1/6',
('PD-1 & CTLA-4', 'Adaptive Lymphoid Cells'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Platelet System'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Granulocyte System'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Define edges to be highlighted in black
def define_black_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
}
# Calculate node positions
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
black_edges = define_black_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
edge_colors = []
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
edge_colors.append('black' if (source, target) in black_edges else 'lightgrey')
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color=edge_colors,
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™ aAPCs", fontsize=18)
plt.show()
# Run the visualization
visualize_nn()
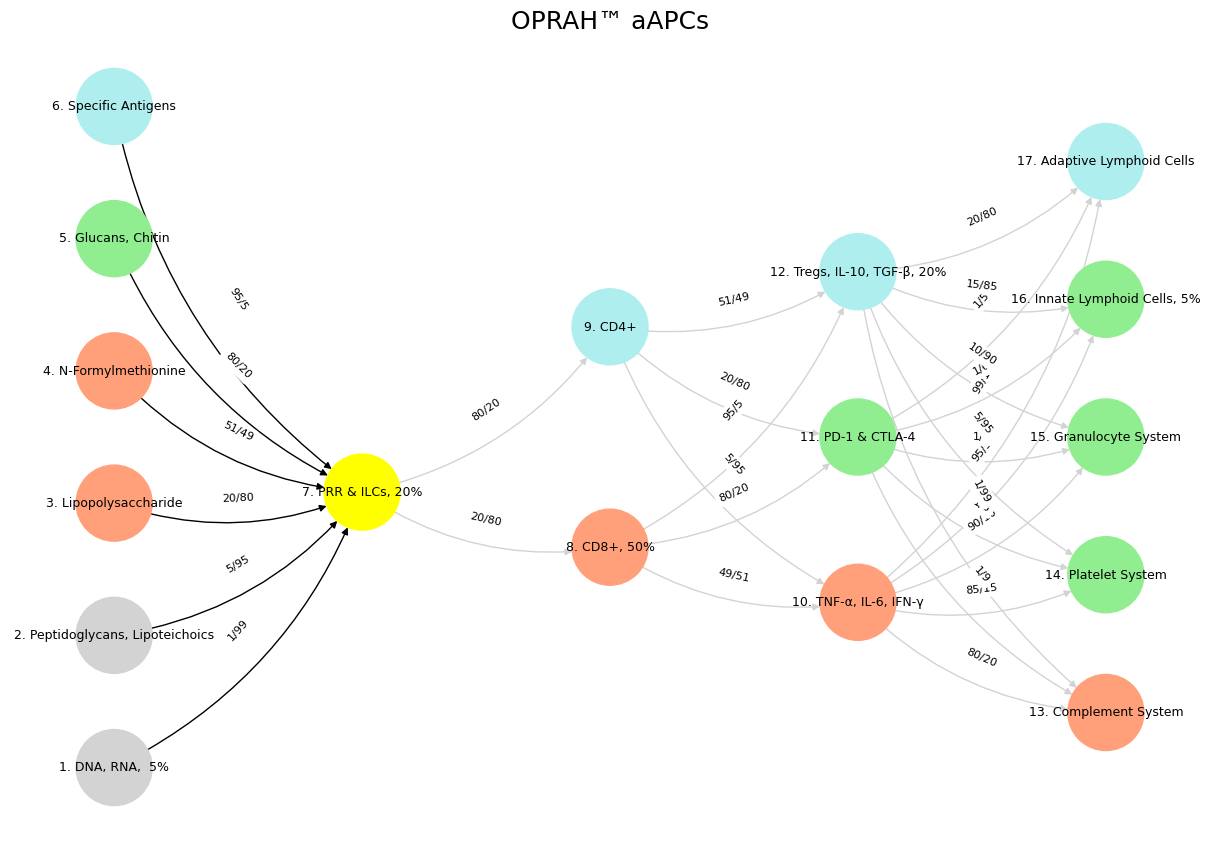
Fig. 23 Nvidia vs. Music. APIs between Nvidias CUDA (server) & their clients (yellowstone node: G1 & G2) are here replaced by the ear-drum (kuhura) & vestibular apparatus (space). The chief enterprise in music is listening and responding (N1, N2, N3) as well as coordination and syncronization with others too (N4 & N5). Whether its classical or improvisational and participatory (time), a massive and infinite combinatorial landscape is available for composer, maestro, performer, audience (rhythm). And who are we to say what exactly music optimizes (semantics)? The mammalian nervous system’s architecture can be mapped onto a neural network and the immune system, revealing a layered interplay of function. Layer 1’s six nodes, embodying cosmic laws and molecular inputs like DNA and antigens, align with the Red Queen Hypothesis, equating sensory and ecosystem via ligand-receptor complementarity. Layer 2, a yellow sentinel node (G1 & G2), parallels MHC and PRRs, relaying perception like cranial and dorsal ganglia. Layer 3’s two decision nodes—instinct (CD8+) versus tempered processes (CD4+)—mirror neural reflexes and modulation. Layer 4’s three equilibrium nodes—self (TNF-α), mutual (PD-1), and other (Tregs)—reflect sympathetic, parasympathetic, and transactional balance. Layer 5’s five metrics (complement to lymphoid cells) optimize responses, akin to neural integration of experience and environment. This mapping highlights a unified biological logic across systems, from static foundations to adaptive outcomes.#

