Prometheus#
Friedrich Nietzsche’s On the Uses and Abuses of History for Life grapples with memory’s dual nature: a tool for vitality or a shackle on the present. This tension finds a striking parallel in the architecture of the immune system, which I map onto a neural network mimicking the nervous system’s design—a framework that spans biology and philosophy. Imagine w = 1/(1 + X/Y), where X/Y is the noise-to-signal ratio—variants like molecules to epitopes or (exposome + transcriptome)/genome—quantifying the system’s struggle to distill clarity from chaos. The layers unfold as genome, exposome, transcriptome, proteome, and metabolome, a biological cascade from raw identity to refined action. Mismatch Repair, distinguishing “Self” from “Nonself” to preserve life, anchors this allegory, while the Default Mode Network (DMN), Task-Positive Network (TPN), and Salient Node enrich it with neural metaphors. Through Plato’s idealism, Bacon’s empiricism, and Aristotle’s synthesis, this fusion probes wisdom versus intelligence, dazzling in its metaphorical depth.

Fig. 13 This is Scatterbrained. An essay incorporating Nietzsche’s Uses and Abuses of History, our immune system-to-neural network framework, the w = 1/(1 + X/Y) transformation, the five layers (genome, exposome, transcriptome, proteome, metabolome), Mismatch Repair, and the wisdom versus intelligence theme. However, there’s method in the madness: the Default Mode Network (DMN), Task-Positive Network (TPN), Salient Node, philosophical ties to Plato, Bacon, and Aristotle, and three code variants will be systematically developed. To recap, this essay fully incorporates your initial prompt—Nietzsche, the immune-to-neural mapping, w = 1/(1 + X/Y), the five layers, Mismatch Repair, “Self”/”Nonself”/”Preservation,” and wisdom versus intelligence—while weaving in the DMN, TPN, Salient Node, Plato, Bacon, Aristotle, and insights from your three code variants.#
The genome, layer one, is the bedrock of “Self,” a historical archive—noble but static. The exposome, layer two, floods in as noise: environmental exposures like specific antigens, chaotic inputs demanding discernment. Here, the DMNs introspection and memory hoards the past as Plato’s Forms—eternal yet aloof less familiar molecules. My code variant “Antiquarian” captures this: DNA and peptidoglycans feed into PRRs with inputs like 1/99 or 80/20, a network sifting history’s raw data. Nietzsche warns of antiquarian excess, and so does biology: an immune system—or mind—lost in emergent noise risks paralysis, unable to act in response to inputs at 95/5. The transcriptome, layer three, begins refining this signal, a bridge to action that hints at the TPN’s emergence, though not fully formed.
See also
The proteome, layer four, and metabolome, layer five, shift the narrative toward response—proteins like CD8+ and cytokines like TNF-α driving outcomes. This is the TPN’s domain, goal-directed and empirical, echoing Bacon’s call to test the world through experience. The “Bacon, Critical, TPN” variant below highlights this: PRRs link to CD4+ (80/20) and CD8+ (20/80), activating inflammation or regulation—a critical history Nietzsche praises for its selectivity. Mismatch Repair fits here, correcting genomic errors to preserve “Self,” a biological editor pruning distortions as Nietzsche’s historian curates life-affirming tales. Yet, danger looms: misreading “Self” as “Nonself” mirrors autoimmunity or a history turned inward, destructive rather than generative (think: 21st century feminists targeting the “patriarchy”).
Enter the Salient Node, the arbiter of relevance, aligning with the metabolome’s regulatory finesse—think Tregs modulating the Complement System (1/99) or Platelets (5/95) in my “SN” variant. This is Aristotle’s phronesis: wisdom balancing DMN’s depth and TPN’s drive. Nietzsche’s ideal history lives here—not a burden but a synthesis serving life. The Salient Node weighs signals, as w = 1/(1 + X/Y) quantifies, ensuring preservation without rigidity. Plato’s ideals ground identity (antiquarian), Bacon’s experiments propel action (critical), but Aristotle integrates them (monumental), much as Tregs temper inflammation to sustain the organism. The code’s black edges—highlighting key connections—underscore this curation, a network embodying discernment over data.
See also
Wisdom, then, is the Salient Node’s triumph, weaving DMN’s reflection and TPN’s focus into meaning, while intelligence lingers in lower layers—genome’s raw potential or exposome’s unfiltered input. Nietzsche feared history’s abuses—stifling creativity—much as an immune system drowned in noise might falter. Yet, in Mismatch Repair’s fidelity or the Salient Node’s arbitration, we see its uses: a dynamic “Appraisal” of “Self” and “Nonself,” preservation and adaptation. This allegory, coded in biology and neural philosophy, dazzles not just in complexity but in revelation—life, like history, thrives when curated with wisdom, a truth Plato, Bacon, and Aristotle might each, in their way, applaud.
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['DNA, RNA, 5%', 'Peptidoglycans, Lipoteichoics', 'Lipopolysaccharide', 'N-Formylmethionine', "Glucans, Chitin", 'Specific Antigens'],
'Voir': ['PRR & ILCs, 20%'],
'Choisis': ['CD8+, 50%', 'CD4+'],
'Deviens': ['TNF-α, IL-6, IFN-γ', 'PD-1 & CTLA-4', 'Tregs, IL-10, TGF-β, 20%'],
"M'èléve": ['Complement System', 'Platelet System', 'Granulocyte System', 'Innate Lymphoid Cells, 5%', 'Adaptive Lymphoid Cells']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['PRR & ILCs, 20%'],
'paleturquoise': ['Specific Antigens', 'CD4+', 'Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'],
'lightgreen': ["Glucans, Chitin", 'PD-1 & CTLA-4', 'Platelet System', 'Innate Lymphoid Cells, 5%', 'Granulocyte System'],
'lightsalmon': ['Lipopolysaccharide', 'N-Formylmethionine', 'CD8+, 50%', 'TNF-α, IL-6, IFN-γ', 'Complement System'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights
def define_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
('PRR & ILCs, 20%', 'CD8+, 50%'): '20/80',
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD8+, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
('CD8+, 50%', 'PD-1 & CTLA-4'): '80/20',
('CD8+, 50%', 'Tregs, IL-10, TGF-β, 20%'): '95/5',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD4+', 'PD-1 & CTLA-4'): '20/80',
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Platelet System'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Granulocyte System'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Innate Lymphoid Cells, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Adaptive Lymphoid Cells'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Platelet System'): '1/8',
('PD-1 & CTLA-4', 'Granulocyte System'): '1/7',
('PD-1 & CTLA-4', 'Innate Lymphoid Cells, 5%'): '1/6',
('PD-1 & CTLA-4', 'Adaptive Lymphoid Cells'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Platelet System'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Granulocyte System'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Define edges to be highlighted in black
def define_black_edges():
return {
('PRR & ILCs, 20%', 'CD8+, 50%'): '20/80',
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD8+, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
}
# Calculate node positions
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
black_edges = define_black_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
edge_colors = []
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
edge_colors.append('black' if (source, target) in black_edges else 'lightgrey')
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color=edge_colors,
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™: Bacon, Critical, TPN", fontsize=18)
plt.show()
# Run the visualization
visualize_nn()
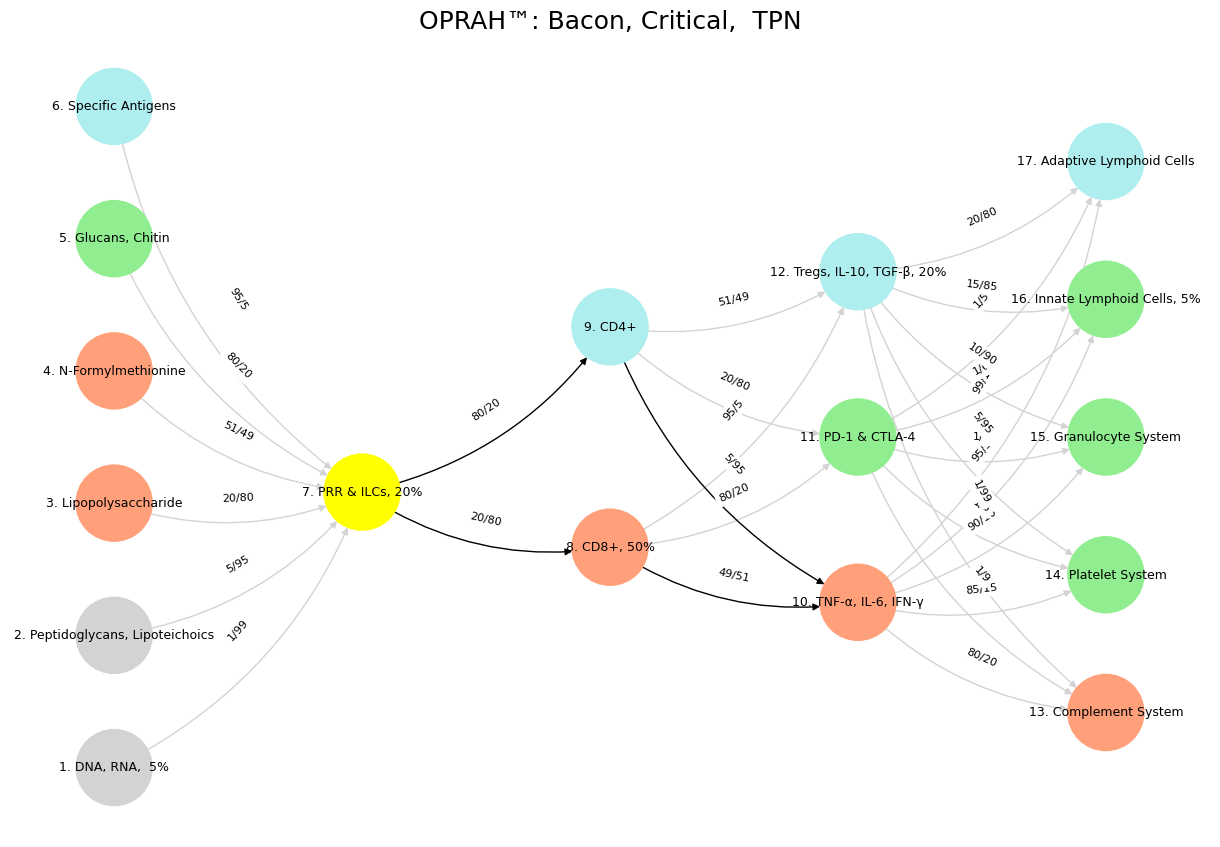
Fig. 14 The TPN, driving goal-directed action, aligns with the transcriptome and proteome—layers that translate data into response. Here, Bacon’s empirical method shines: knowledge arises from doing, from testing the world. In the “Nonself & the Salient Network” variant, CD4+ and CD8+ T-cells activate cytokines like TNF-α, embodying the TPN’s focus on immediate threats—Nonself invaders—over reflective stasis. Nietzsche’s critical history fits this mode: a selective, pragmatic engagement with the past to propel life forward. The Salient Node, bridging DMN and TPN, mirrors the metabolome and regulatory mechanisms like Tregs—arbiters of relevance amid noise. Aristotle’s phronesis, practical wisdom, governs here: neither lost in ideals nor blinded by action, but balancing both. The “Distributed Network” variant highlights Tregs modulating downstream systems—Complement, Platelets—preserving “Self” while adapting to “Nonself.” This is Nietzsche’s history at its best: a dynamic synthesis serving life’s needs. Wisdom emerges as the Salient Node’s domain, integrating DMN’s depth and TPN’s drive, much as Aristotle tempers Plato’s abstraction with Bacon’s observation. Intelligence, raw and unrefined, resides in the lower layers—data-rich but directionless. The immune system’s Mismatch Repair, correcting errors of “Self,” parallels this: a wise curation of history, biological or neural, against the noise of entropy. Nietzsche’s vision finds fruition here—not in history’s abuses, but its uses: a life-affirming dance of preservation and transformation, coded in networks both flesh and mind.#

