Dancing in Chains#
The integration of thalamocortical gating under N2 refines the neural network’s function by explicitly accounting for the regulatory mechanisms that dictate sensory flow to the cortex. The thalamus, positioned as an essential relay hub, serves as both a gatekeeper and a modulator of ascending sensory input before it reaches higher cortical processing. Encoding this under N2 aligns with its role in dynamically filtering external stimuli, ensuring that relevant signals pass while extraneous noise is suppressed. This process is fundamental for maintaining equilibrium between attentional control and cognitive flexibility, reinforcing the dynamic capability of the network.

Fig. 34 Freedom in Fetters—a Princely Freedom. Chopin, the last of the modern musicians, who gazed at and worshipped beauty, like Leopardi; Chopin, the Pole, the inimitable (none that came before or after him has a right to this name)—Chopin had the same princely punctilio in convention (grammar, space) that Raphael shows in the use of the simplest traditional colours. The only difference is that Chopin applies them not to colour but to melodic and rhythmic traditions (prosody, time). He admitted the validity of these traditions because he was born under the sway of etiquette. But in these fetters he plays and dances as the freest and daintiest of spirits, and, be it observed, he does not spurn the chain. Source: Human All-Too-Human Part II#
Within this updated structure, the parasympathetic (feed, breed, sleep) and sympathetic (fight, flight, fright) axes represent survival-based dynamic capability, each modulating responses through distinct neuromodulatory pathways. The parasympathetic system, characterized by acetylcholine-dominated homeostasis, facilitates functions that prioritize resource conservation, social bonding, and long-term biological stability. In contrast, the sympathetic system, largely governed by norepinephrine and stress responses, drives immediate, high-stakes decision-making under pressure. These systems interface through G3, the presynaptic autonomic ganglia, which serves as the transactional jostling node, shifting activation between homeostatic maintenance and high-adrenaline mobilization.
The Salient Network influences the noise-signal ratio
– Yours Truly
This dichotomy directly maps onto computational principles found in reinforcement learning, where feed, breed, sleep parallels an exploitation strategy—optimizing existing resources—while fight, flight, fright mirrors exploration, which prioritizes rapid adaptation to novel threats. The inclusion of thalamocortical gating under N2 further sharpens this analogy by embedding a regulatory feedback mechanism, allowing the system to modulate the balance between focused engagement and inhibition. This reinforces N1-N3 (ascending activators) as primary conduits of agentic decision-making while positioning N4-N5 (descending inhibitors) as essential controls that prevent maladaptive hyperactivity.
The relationship between the default mode network (DMN) and parasympathetic activity warrants interrogation within this framework. The DMN, primarily active during introspective and self-referential thought, is often contrasted with task-positive networks that drive active engagement with the external world. Given that parasympathetic states prioritize rest, memory consolidation, and social attachment, it would seem intuitive to align the DMN with parasympathetic dominance. However, the complexity arises from thalamocortical gating, which dictates the degree of cortical involvement. If the DMN is understood as a form of predictive modeling rather than pure rest, its function transcends mere parasympathetic alignment and enters a meta-regulatory domain that integrates both sympathetic and parasympathetic modulations.
Thus, while the DMN’s introspective nature overlaps with parasympathetic functions such as memory consolidation (sleep) and social cognition (breed), it remains insufficiently defined by purely parasympathetic characteristics. Instead, it operates at an emergent level of cognitive homeostasis, incorporating inputs from both the sympathetic-driven alertness system and the parasympathetic-driven restoration process. This suggests that rather than existing as a direct analog to parasympathetic activity, the DMN should be seen as a higher-order regulator that modulates the interplay between dynamic capabilities.
By encoding thalamocortical gating under N2, this neural network model now captures an essential mechanism for sensory and cognitive regulation. The incorporation of sympathetic-parasympathetic duality, dynamic gating, and computational reinforcement principles allows for a more granular representation of agentic capability. The DMN, rather than being reducible to a singular autonomic mode, represents a higher-order adaptive network, integrating parasympathetic recuperation with the strategic anticipation required for navigating complex social and environmental landscapes.
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['Genome, 5%', 'Culture', 'Nourish It', 'Know It', "Move It", 'Injure It'], # Static
'Voir': ['Exposome, 15%'],
'Choisis': ['Metabolome, 50%', 'Basal Metabolic Rate'],
'Deviens': ['Unstructured-Intense', 'Weekly-Calendar', 'Proteome, 25%'],
"M'èléve": ['NexToken Prediction', 'Hydration', 'Fat-Muscle Ratio', 'Amor Fatì, 5%', 'Existential Cadence']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['Exposome, 15%'],
'paleturquoise': ['Injure It', 'Basal Metabolic Rate', 'Proteome, 25%', 'Existential Cadence'],
'lightgreen': ["Move It", 'Weekly-Calendar', 'Hydration', 'Amor Fatì, 5%', 'Fat-Muscle Ratio'],
'lightsalmon': ['Nourish It', 'Know It', 'Metabolome, 50%', 'Unstructured-Intense', 'NexToken Prediction'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights (hardcoded for editing)
def define_edges():
return {
('Genome, 5%', 'Exposome, 15%'): '1/99',
('Culture', 'Exposome, 15%'): '5/95',
('Nourish It', 'Exposome, 15%'): '20/80',
('Know It', 'Exposome, 15%'): '51/49',
("Move It", 'Exposome, 15%'): '80/20',
('Injure It', 'Exposome, 15%'): '95/5',
('Exposome, 15%', 'Metabolome, 50%'): '20/80',
('Exposome, 15%', 'Basal Metabolic Rate'): '80/20',
('Metabolome, 50%', 'Unstructured-Intense'): '49/51',
('Metabolome, 50%', 'Weekly-Calendar'): '80/20',
('Metabolome, 50%', 'Proteome, 25%'): '95/5',
('Basal Metabolic Rate', 'Unstructured-Intense'): '5/95',
('Basal Metabolic Rate', 'Weekly-Calendar'): '20/80',
('Basal Metabolic Rate', 'Proteome, 25%'): '51/49',
('Unstructured-Intense', 'NexToken Prediction'): '80/20',
('Unstructured-Intense', 'Hydration'): '85/15',
('Unstructured-Intense', 'Fat-Muscle Ratio'): '90/10',
('Unstructured-Intense', 'Amor Fatì, 5%'): '95/5',
('Unstructured-Intense', 'Existential Cadence'): '99/1',
('Weekly-Calendar', 'NexToken Prediction'): '1/9',
('Weekly-Calendar', 'Hydration'): '1/8',
('Weekly-Calendar', 'Fat-Muscle Ratio'): '1/7',
('Weekly-Calendar', 'Amor Fatì, 5%'): '1/6',
('Weekly-Calendar', 'Existential Cadence'): '1/5',
('Proteome, 25%', 'NexToken Prediction'): '1/99',
('Proteome, 25%', 'Hydration'): '5/95',
('Proteome, 25%', 'Fat-Muscle Ratio'): '10/90',
('Proteome, 25%', 'Amor Fatì, 5%'): '15/85',
('Proteome, 25%', 'Existential Cadence'): '20/80'
}
# Calculate positions for nodes
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color='gray',
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™: Heredity, Lifestyle, Badluck", fontsize=25)
plt.show()
# Run the visualization
visualize_nn()
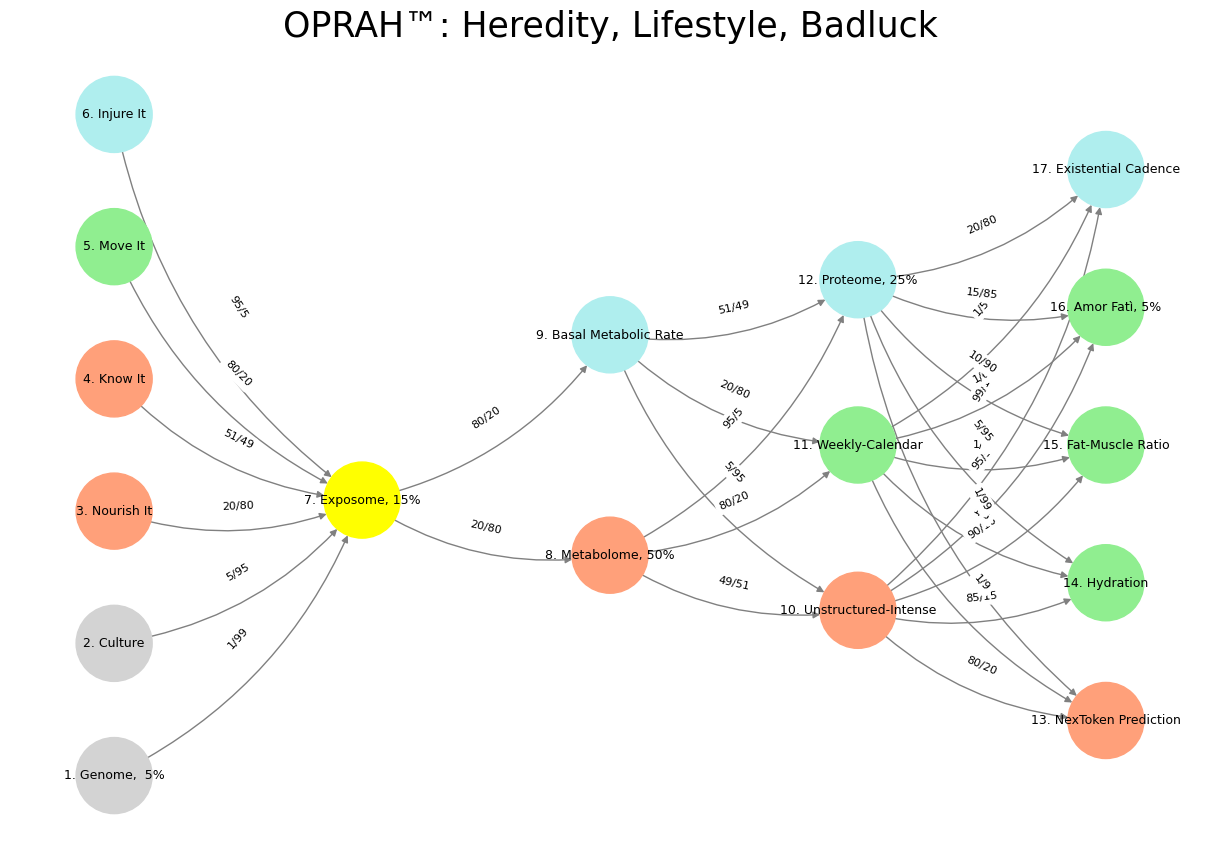

Fig. 35 G1-G3: Ganglia & N1-N5 Nuclei. These are cranial nerve, dorsal-root (G1 & G2); basal ganglia, thalamus, hypothalamus (N1, N2, N3); and brain stem and cerebelum (N4 & N5).#

