#
1#
Show code cell source
suppressWarnings({
invisible({
rm(list = ls())
if (!require(icdpicr, quietly = TRUE, warn.conflicts = FALSE)) install.packages('icdpicr')
if (!require(dplyr, quietly = TRUE, warn.conflicts = FALSE)) install.packages('dplyr')
if (!require(readr, quietly = TRUE, warn.conflicts = FALSE)) install.packages('readr')
if (!require(tidyr, quietly = TRUE, warn.conflicts = FALSE)) install.packages('tidyr')
if (!require(ggplot2, quietly = TRUE, warn.conflicts = FALSE)) install.packages('ggplot2')
if (!require(gganimate, quietly = TRUE, warn.conflicts = FALSE)) install.packages('gganimate')
if (!require(magick, quietly = TRUE, warn.conflicts = FALSE)) install.packages('magick')
if (!require(gifski, quietly = TRUE, warn.conflicts = FALSE)) install.packages('gifski')
library(icdpicr, quietly = TRUE, warn.conflicts = FALSE)
library(dplyr, quietly = TRUE, warn.conflicts = FALSE)
library(readr, quietly = TRUE, warn.conflicts = FALSE)
library(tidyr, quietly = TRUE, warn.conflicts = FALSE)
library(ggplot2, quietly = TRUE, warn.conflicts = FALSE)
library(gganimate, quietly = TRUE, warn.conflicts = FALSE)
library(magick, quietly = TRUE, warn.conflicts = FALSE)
library(gifski, quietly = TRUE, warn.conflicts = FALSE)
})
})
The downloaded binary packages are in
/var/folders/sx/fd6zgj191mx45hspzbgwzlnr0000gn/T//RtmpuceRvB/downloaded_packages
2#
Show code cell source
# Loading required libraries
library(ggplot2)
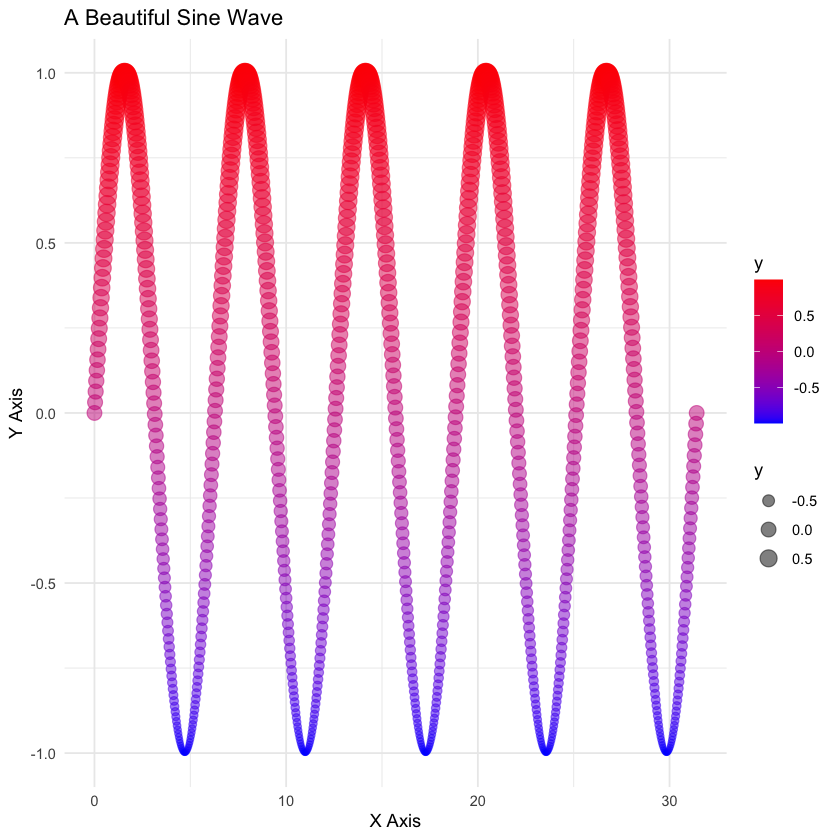
# Creating a data frame with 1,000 points following a sine wave
n_points <- 1000
data <- data.frame(
x = seq(0, 10 * pi, length.out = n_points),
y = sin(seq(0, 10 * pi, length.out = n_points))
)
# Using ggplot2 to create the plot
ggplot(data, aes(x = x, y = y, color = y, size = y)) +
geom_point(alpha = 0.5) +
scale_color_gradient(low = "blue", high = "red") +
scale_size_continuous(range = c(1, 5)) +
theme_minimal() +
labs(title = "A Beautiful Sine Wave",
x = "X Axis",
y = "Y Axis")
3#
Show code cell source
fibonacci <- local({
memo <- numeric()
function(n) {
if (n <= 1) return(n)
if (!is.na(memo[n])) return(memo[n])
memo[n] <<- fibonacci(n - 1) + fibonacci(n - 2)
return(memo[n])
}
})
# Example usage
n <- 10
cat("The first", n, "numbers in the Fibonacci sequence are:")
sapply(0:(n-1), fibonacci)
The first 10 numbers in the Fibonacci sequence are:
- 0
- 1
- 1
- 2
- 3
- 5
- 8
- 13
- 21
- 34
4#
Show code cell source
# Loading libraries
library(ggplot2)
library(gganimate)
# Defining parameters for a particle in a magnetic field
t <- seq(0, 10, by = 0.1)
x <- sin(t)
y <- cos(t)
# Creating a data frame to hold the values
particle_data <- data.frame(
time = t,
x = x,
y = y
)
# Creating the plot using ggplot2 and gganimate
plot <- ggplot(particle_data, aes(x = x, y = y)) +
geom_path(aes(group = 1), color = 'gray') + # Making sure all points are in the same group
geom_point(size = 4) +
labs(title = "Particle in a Magnetic Field",
x = "X Axis",
y = "Y Axis") +
theme_minimal() +
coord_fixed(ratio = 1) +
transition_reveal(time)
# Create and save the animation
animate(plot, renderer = gifski_renderer("particle_animation.gif"))
`geom_path()`: Each group consists of only one observation.
i Do you need to adjust the group aesthetic?
`geom_path()`: Each group consists of only one observation.
i Do you need to adjust the group aesthetic?