Veiled Resentment#

Uddin: a Kindred Spirit. Our gh-pages based ecosystem integration & navigation (EIN) framework is a competitive solution to a diagnosis we reached independently of Uddin. Source: Draft Complaint
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['The Great York, 5%', 'Peptidoglycans, Lipoteichoics', 'Lipopolysaccharide', 'N-Formylmethionine', "Glucans, Chitin", 'Specific Antigens'],
'Voir': ['Empire Unpossesed, 20%'],
'Choisis': ['Yorks Heirs Alive, 50%', 'King of England'],
'Deviens': ['Sword Unswayed', 'Chair Empty', 'King Dead, 20%'],
"M'èléve": ['Why Then at Sea?', 'Platelet System', 'Granulocyte System', 'Innate Lymphoid Cells, 5%', 'Adaptive Lymphoid Cells']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['Empire Unpossesed, 20%'],
'paleturquoise': ['Specific Antigens', 'King of England', 'King Dead, 20%', 'Adaptive Lymphoid Cells'],
'lightgreen': ["Glucans, Chitin", 'Chair Empty', 'Platelet System', 'Innate Lymphoid Cells, 5%', 'Granulocyte System'],
'lightsalmon': ['Lipopolysaccharide', 'N-Formylmethionine', 'Yorks Heirs Alive, 50%', 'Sword Unswayed', 'Why Then at Sea?'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights
def define_edges():
return {
('The Great York, 5%', 'Empire Unpossesed, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'Empire Unpossesed, 20%'): '5/95',
('Lipopolysaccharide', 'Empire Unpossesed, 20%'): '20/80',
('N-Formylmethionine', 'Empire Unpossesed, 20%'): '51/49',
("Glucans, Chitin", 'Empire Unpossesed, 20%'): '80/20',
('Specific Antigens', 'Empire Unpossesed, 20%'): '95/5',
('Empire Unpossesed, 20%', 'Yorks Heirs Alive, 50%'): '20/80',
('Empire Unpossesed, 20%', 'King of England'): '80/20',
('Yorks Heirs Alive, 50%', 'Sword Unswayed'): '49/51',
('Yorks Heirs Alive, 50%', 'Chair Empty'): '80/20',
('Yorks Heirs Alive, 50%', 'King Dead, 20%'): '95/5',
('King of England', 'Sword Unswayed'): '5/95',
('King of England', 'Chair Empty'): '20/80',
('King of England', 'King Dead, 20%'): '51/49',
('Sword Unswayed', 'Why Then at Sea?'): '80/20',
('Sword Unswayed', 'Platelet System'): '85/15',
('Sword Unswayed', 'Granulocyte System'): '90/10',
('Sword Unswayed', 'Innate Lymphoid Cells, 5%'): '95/5',
('Sword Unswayed', 'Adaptive Lymphoid Cells'): '99/1',
('Chair Empty', 'Why Then at Sea?'): '1/9',
('Chair Empty', 'Platelet System'): '1/8',
('Chair Empty', 'Granulocyte System'): '1/7',
('Chair Empty', 'Innate Lymphoid Cells, 5%'): '1/6',
('Chair Empty', 'Adaptive Lymphoid Cells'): '1/5',
('King Dead, 20%', 'Why Then at Sea?'): '1/99',
('King Dead, 20%', 'Platelet System'): '5/95',
('King Dead, 20%', 'Granulocyte System'): '10/90',
('King Dead, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('King Dead, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Define edges to be highlighted in black
def define_black_edges():
return {
('King Dead, 20%', 'Why Then at Sea?'): '1/99',
('King Dead, 20%', 'Platelet System'): '5/95',
('King Dead, 20%', 'Granulocyte System'): '10/90',
('King Dead, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('King Dead, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Calculate node positions
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
black_edges = define_black_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
edge_colors = []
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
edge_colors.append('black' if (source, target) in black_edges else 'lightgrey')
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color=edge_colors,
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™: Richard III", fontsize=18)
plt.show()
# Run the visualization
visualize_nn()
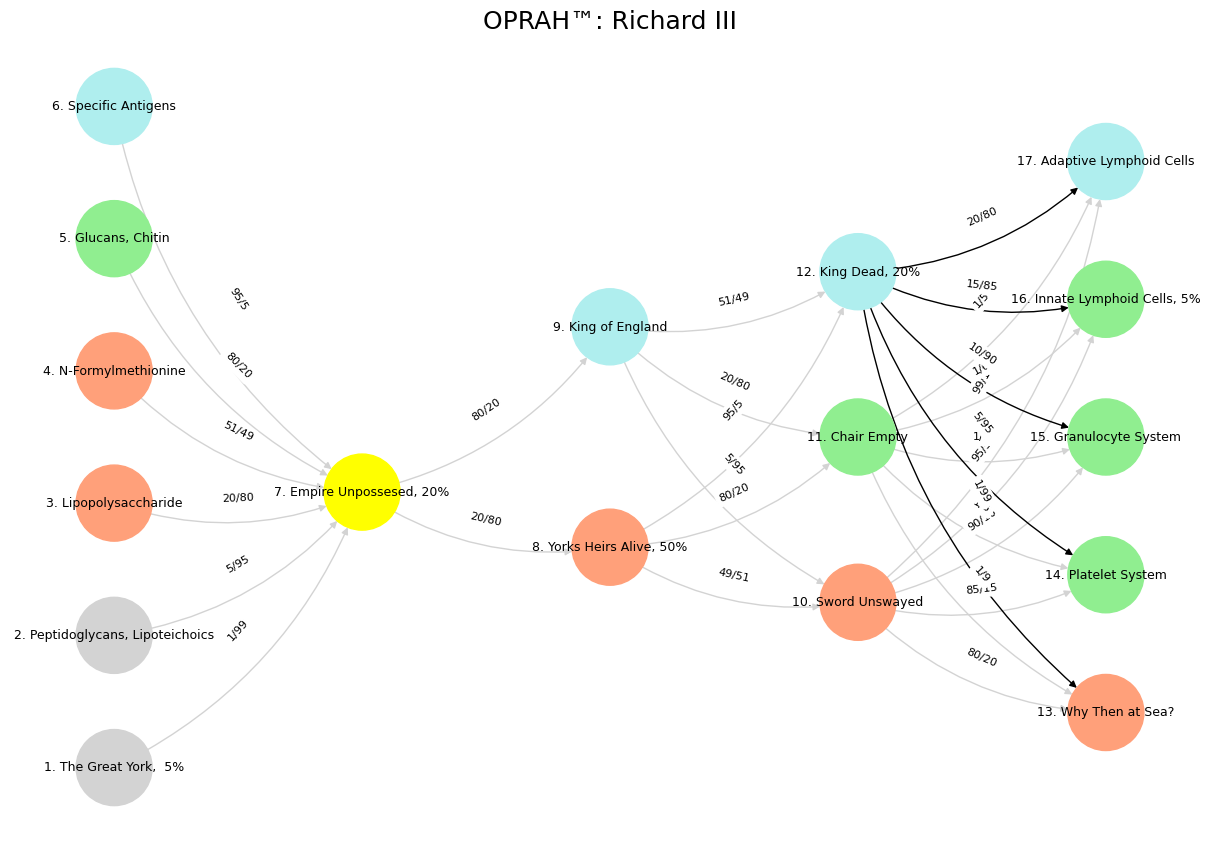
Fig. 15 Is the chair empty? Is the sword unswayed? Is the King dead? The empire unpossessed? What heir of York is there alive but we? And who is England’s King but great York’s heir? Then tell me, what makes he upon the seas? # Our Bequest: An Epiloue: In the dying light of distant stars, we leave our mark—a brief flicker in cosmic time. Our story is written in layers: In cosmology: We are stardust awakened, conscious matter that gazed back at its origins, mapping the very forces that birthed us, reaching toward comprehension of a universe destined for cold expansion. In geology: We etched our presence into stone, leaving behind the Anthropocene—a thin stratum of plastic, concrete, and radiation that will outlast our memory by millions of years. In biology: We carried forward the unbroken chain of life, reshaping genomes while ourselves remaining vessels of the ancient code that stretches back to the first cell. In ecology: We wove ourselves into and then partially unwove the intricate tapestry of Earth’s living systems, becoming both keystone and burden to countless species with whom we shared this planetary moment. In symbiotology: We discovered our true nature as colonies of cooperation—from the mitochondria within our cells to the microbial gardens we cultivate and are cultivated by—revealing that boundaries between self and other were never as firm as we imagined. In teleology: We sought purpose in a universe that offered none, creating meaning through consciousness, art, love, and wonder—transforming blind causality into narrative, accident into significance. Our legacy is this paradox: supremely powerful yet fundamentally fragile, capable of both destruction and creation, ultimately transient yet eternally connected to what came before and what follows after.#

