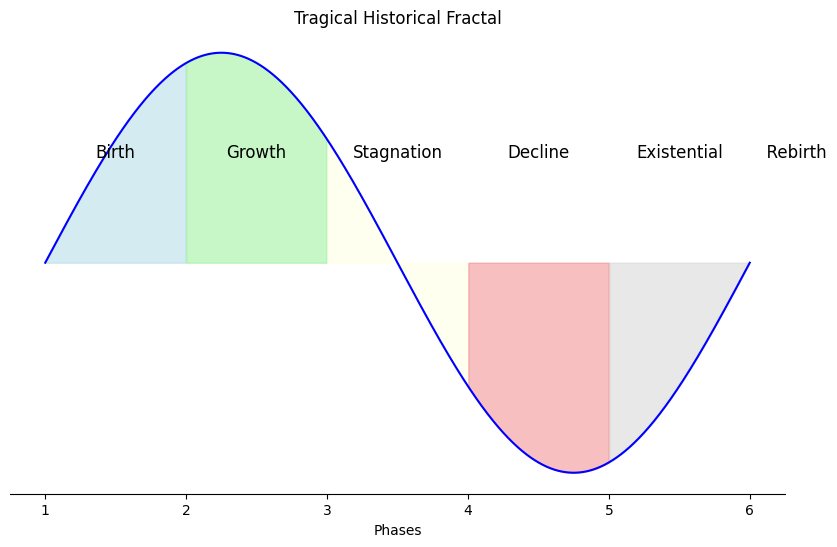
Industry#
Finance, electrons#
Last summer in Seattle, I was reduced to an electron zapping around a circuit-board 4
1. Phonetics
\
2. Temperament -> 4. Modes -> 5. NexToken -> 6. Emotion
/
3. Scales
Show code cell source
import networkx as nx
import matplotlib.pyplot as plt
def add_fern_edges(G, start, depth, angle, scale):
if depth == 0:
return
end = (start[0] + scale * angle[0], start[1] + scale * angle[1])
G.add_edge(start, end)
new_angle1 = (angle[0] * 0.6 - angle[1] * 0.4, angle[0] * 0.4 + angle[1] * 0.6)
new_angle2 = (angle[0] * 0.6 + angle[1] * 0.4, -angle[0] * 0.4 + angle[1] * 0.6)
add_fern_edges(G, end, depth - 1, new_angle1, scale * 0.7)
add_fern_edges(G, end, depth - 1, new_angle2, scale * 0.7)
def create_fern_graph(depth, scale=1):
G = nx.Graph()
start = (0, 0)
angle = (0, 1)
add_fern_edges(G, start, depth, angle, scale)
return G
# Generate the fern graph
fern_depth = 7 # Adjust depth for more or fewer branches
fern_graph = create_fern_graph(fern_depth)
# Define colors
leaf_color = '#5F9E54' # A green between lime and cabbage
stem_color = '#3B6631' # A darker green for the stem
# Visualize the fern
plt.figure(figsize=(8, 12))
# Draw edges with different colors
edges = fern_graph.edges()
colors = [stem_color if i == 0 else leaf_color for i, (u, v) in enumerate(edges)]
pos = {node: node for node in fern_graph.nodes()}
nx.draw(fern_graph, pos, with_labels=False, node_size=0, edge_color=colors, width=2)
plt.axis('off')
# Save the image
# plt.savefig("/Users/apollo/Documents/rhythm/music/kitabo/ensi/figures/fern_fractal.png", dpi=300, bbox_inches='tight')
plt.show()
Show code cell output
Show code cell source
import networkx as nx
import matplotlib.pyplot as plt
import numpy as np
# Create a directed graph
G = nx.DiGraph()
# Add nodes representing different levels (subatomic, atomic, cosmic, financial, social)
levels = ['Cosmic', 'Subatomic', 'Atomic', 'Molecular', 'Biochemical', 'Financial', 'Social']
# Add nodes to the graph
G.add_nodes_from(levels)
# Add edges to represent the flow of information (photons)
# Assuming the flow is directional from more fundamental levels to more complex ones
edges = [('Cosmic', 'Subatomic'),
('Subatomic', 'Atomic'),
('Atomic', 'Molecular'),
('Molecular', 'Biochemical'),
('Biochemical', 'Financial'),
('Financial', 'Social')]
# Add edges to the graph
G.add_edges_from(edges)
# Define positions for the nodes in a circular layout
pos = nx.circular_layout(G)
# Set the figure size (width, height)
plt.figure(figsize=(10, 10)) # Adjust the size as needed
# Draw the main nodes
nx.draw_networkx_nodes(G, pos, node_color='lightblue', node_size=3000)
# Draw the edges with arrows and create space between the arrowhead and the node
nx.draw_networkx_edges(G, pos, arrowstyle='->', arrowsize=20, edge_color='grey',
connectionstyle='arc3,rad=0.2') # Adjust rad for more/less space
# Add smaller red nodes (photon nodes) exactly on the circular layout
for edge in edges:
# Calculate the vector between the two nodes
vector = pos[edge[1]] - pos[edge[0]]
# Calculate the midpoint
mid_point = pos[edge[0]] + 0.5 * vector
# Normalize to ensure it's on the circle
radius = np.linalg.norm(pos[edge[0]])
mid_point_on_circle = mid_point / np.linalg.norm(mid_point) * radius
# Draw the small red photon node at the midpoint on the circular layout
plt.scatter(mid_point_on_circle[0], mid_point_on_circle[1], c='lightpink', s=500, zorder=3)
# Draw a small lime green arrow inside the red node to indicate direction
arrow_vector = vector / np.linalg.norm(vector) * 0.1 # Scale down arrow size
plt.arrow(mid_point_on_circle[0] - 0.05 * arrow_vector[0],
mid_point_on_circle[1] - 0.05 * arrow_vector[1],
arrow_vector[0], arrow_vector[1],
head_width=0.03, head_length=0.05, fc='limegreen', ec='limegreen', zorder=4)
# Draw the labels for the main nodes
nx.draw_networkx_labels(G, pos, font_size=9, font_weight='normal')
# Add a legend for "Photon/Info"
plt.scatter([], [], c='lightpink', s=100, label='Photon/Info') # Empty scatter for the legend
plt.legend(scatterpoints=1, frameon=True, labelspacing=1, loc='upper right')
# Set the title and display the plot
plt.title('Flow of Information Across Levels', fontsize=15)
plt.axis('off')
plt.show()
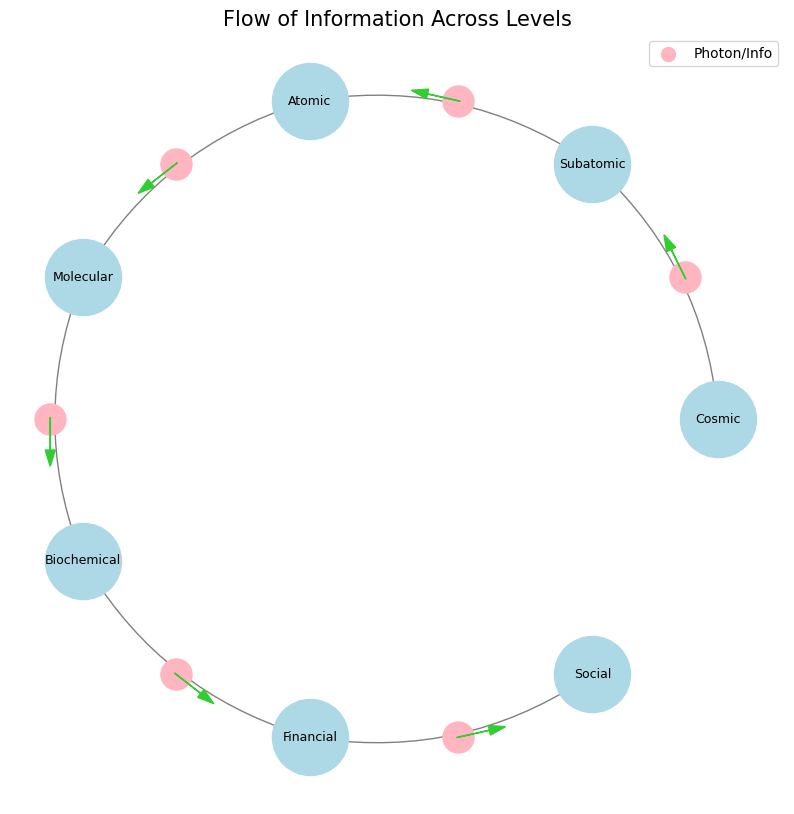

Information at core is photons. It’s electrons, or electrons changing energy, which is photons, and that’s processes. So, from one node via a wedge to another node, things move at the speed of light. They may not seem that way, but they do. So, in physics, subatomical, self-explanatory, you know, an electron moves from a high energy level to a low energy level, gives a photon, and blah, blah, blah, blah. Processes are set into play. At the atomic level and molecular level, that could lead to chemical reactions. Well, the human senses could detect changes in color, or emission of rays, and all that. So, there’s information. There’s information flow, and it’s photons. When you go to the cosmic level, you know, whatever is causing, you know, the cosmos to rotate around certain centers as it expands, in general, but in specific locales, like in, you know, like one galaxy, this stuff rotating around some mystical thing. The mystical thing is transmitting information more than anything else, and that’s the center. That’s like a leader. That’s gravity. Now, while the force there may not be a photon, it is something in the zone of the speed of light. In finance, when markets respond to information in microseconds, you know, it’s because, again, of photons via electronics, electronic media, and electronic devices. So, back to photons, and electrons, and information, nodes, and wedges. So, from the physical to the biological, the biochemistry, and to the social, it’s photons. It’s information. It’s nodes and edges. One step to the next step in the process. That arrow of the process is a photon. Could you give me some code that visualizes this again? What you’re describing is an intricate visualization of information flow across different scales—from subatomic particles to cosmic structures, finance, and social networks—using the concept of photons as carriers of information. This visualization would essentially map nodes (points of information) and edges (connections or flows of information) with arrows representing the directional flow of photons, or in a broader sense, the transfer of energy or information. Here’s some Python code using NetworkX and Matplotlib to create a basic visualization of this concept: Explanation. Nodes represent different levels of complexity, from the subatomic to the social. Edges represent the flow of information, symbolized by photons or analogous concepts, moving from one level to the next. The circular layout helps to visualize these connections in a more interconnected way, reflecting how these processes are not linear but cyclical or networked. Visualization Concept: This code creates a directed graph where each node represents a different scale of information processing or complexity, and each edge represents the flow of information (or photons) between these levels. The graph is set up to highlight the idea that information flows through various levels, from the most fundamental (subatomic) to the most complex (social networks), with photons or analogous concepts acting as the arrows of information transfer. Dead-space between Social & Cosmic is occupied by StarTrek, StarWars, and anything human or AI that can leave a mark on the cosmos. Also, the only perceptilble deceleration in info-transfer comes after “Biochemical”, and represents “Free-Will” or prevarication (Inspired by Industry) 55#
ii \(\mu\) Fractals, God, \(N\)#
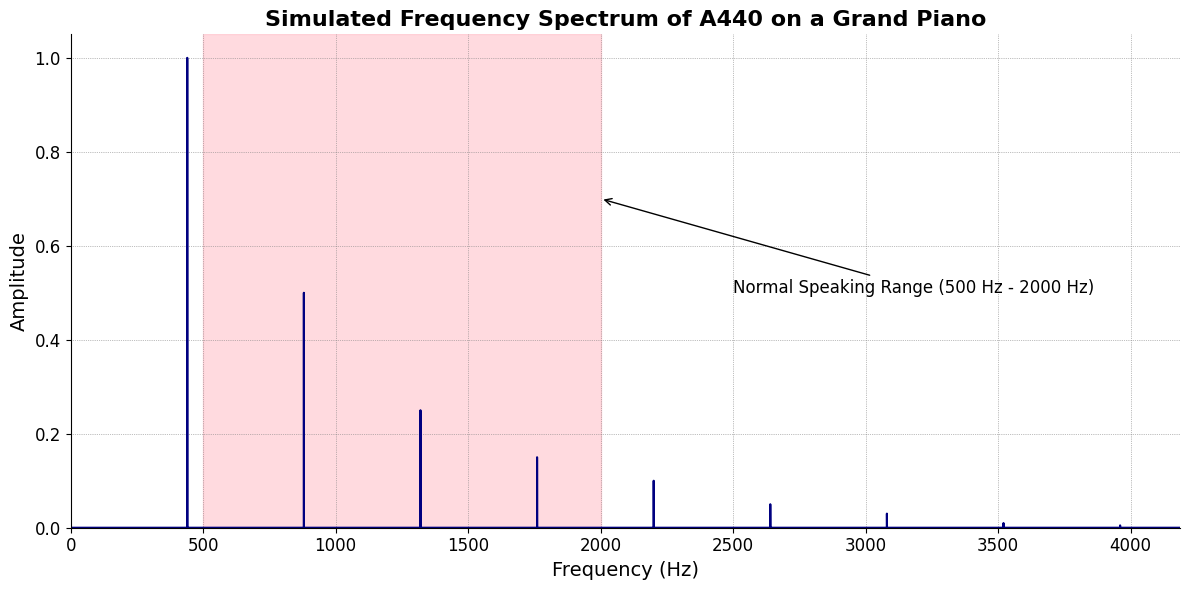
ii\(f(t)\) Phonetics:Fractals\(440Hz \times 2^{\frac{N}{12}}\), \(S_0(t) \times e^{logHR}\), \(\frac{S N(d_1)}{K N(d_2)} \times e^{rT}\)
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
# Parameters
sample_rate = 44100 # Hz
duration = 20.0 # seconds
A4_freq = 440.0 # Hz
# Time array
t = np.linspace(0, duration, int(sample_rate * duration), endpoint=False)
# Fundamental frequency (A4)
signal = np.sin(2 * np.pi * A4_freq * t)
# Adding overtones (harmonics)
harmonics = [2, 3, 4, 5, 6, 7, 8, 9] # First few harmonics
amplitudes = [0.5, 0.25, 0.15, 0.1, 0.05, 0.03, 0.01, 0.005] # Amplitudes for each harmonic
for i, harmonic in enumerate(harmonics):
signal += amplitudes[i] * np.sin(2 * np.pi * A4_freq * harmonic * t)
# Perform FFT (Fast Fourier Transform)
N = len(signal)
yf = np.fft.fft(signal)
xf = np.fft.fftfreq(N, 1 / sample_rate)
# Plot the frequency spectrum
plt.figure(figsize=(12, 6))
plt.plot(xf[:N//2], 2.0/N * np.abs(yf[:N//2]), color='navy', lw=1.5)
# Aesthetics improvements
plt.title('Simulated Frequency Spectrum of A440 on a Grand Piano', fontsize=16, weight='bold')
plt.xlabel('Frequency (Hz)', fontsize=14)
plt.ylabel('Amplitude', fontsize=14)
plt.xlim(0, 4186) # Limit to the highest frequency on a piano (C8)
plt.ylim(0, None)
# Shading the region for normal speaking range (approximately 85 Hz to 255 Hz)
plt.axvspan(500, 2000, color='lightpink', alpha=0.5)
# Annotate the shaded region
plt.annotate('Normal Speaking Range (500 Hz - 2000 Hz)',
xy=(2000, 0.7), xycoords='data',
xytext=(2500, 0.5), textcoords='data',
arrowprops=dict(facecolor='black', arrowstyle="->"),
fontsize=12, color='black')
# Remove top and right spines
plt.gca().spines['top'].set_visible(False)
plt.gca().spines['right'].set_visible(False)
# Customize ticks
plt.xticks(fontsize=12)
plt.yticks(fontsize=12)
# Light grid
plt.grid(color='grey', linestyle=':', linewidth=0.5)
# Show the plot
plt.tight_layout()
plt.show()
Show code cell output
V7\(S(t)\) Temperament:ChainsEqual temperament, Proportional hazards, Homoskedastic volatilityi\(h(t)\) Scale:Equations9 Inherited, Added, Overcome
V7 \(\sigma\) Chains, Muna, \(\beta\)#
\((X'X)^T \cdot X'Y\): Mode: \( \mathcal{F}(t) = \alpha \cdot \left( \prod_{i=1}^{n} \frac{\partial \psi_i(t)}{\partial t} \right) + \beta \cdot \int_{0}^{t} \left( \sum_{j=1}^{m} \frac{\partial \phi_j(\tau)}{\partial \tau} \right) d\tau\) .
Accidentsmezcal, mezclar, mezcaline. Just a reminder that accidents can mislead one to perceive patterns where there’s no underlying process responsible for them. That said, \(\alpha\) & \(beta\) are emerging as parameters representing the highest hierarchy in this multilevel dataset. They stand for determined (chains) & freewill (wiggle-room)
Show code cell source
import matplotlib.pyplot as plt
import numpy as np
# Clock settings; f(t) random disturbances making "paradise lost"
clock_face_radius = 1.0
number_of_ticks = 7
tick_labels = [
"Root-iADL (i)",
"Hunter-gather (ii7♭5)", "Peasant (III)", "Farmer (iv)", "Manufacturer (V7♭9♯9♭13)",
"Energy (VI)", "Transport (VII)"
]
# Calculate the angles for each tick (in radians)
angles = np.linspace(0, 2 * np.pi, number_of_ticks, endpoint=False)
# Inverting the order to make it counterclockwise
angles = angles[::-1]
# Create figure and axis
fig, ax = plt.subplots(figsize=(8, 8))
ax.set_xlim(-1.2, 1.2)
ax.set_ylim(-1.2, 1.2)
ax.set_aspect('equal')
# Draw the clock face
clock_face = plt.Circle((0, 0), clock_face_radius, color='lightgrey', fill=True)
ax.add_patch(clock_face)
# Draw the ticks and labels
for angle, label in zip(angles, tick_labels):
x = clock_face_radius * np.cos(angle)
y = clock_face_radius * np.sin(angle)
# Draw the tick
ax.plot([0, x], [0, y], color='black')
# Positioning the labels slightly outside the clock face
label_x = 1.1 * clock_face_radius * np.cos(angle)
label_y = 1.1 * clock_face_radius * np.sin(angle)
# Adjusting label alignment based on its position
ha = 'center'
va = 'center'
if np.cos(angle) > 0:
ha = 'left'
elif np.cos(angle) < 0:
ha = 'right'
if np.sin(angle) > 0:
va = 'bottom'
elif np.sin(angle) < 0:
va = 'top'
ax.text(label_x, label_y, label, horizontalalignment=ha, verticalalignment=va, fontsize=10)
# Remove axes
ax.axis('off')
# Show the plot
plt.show()
Show code cell output
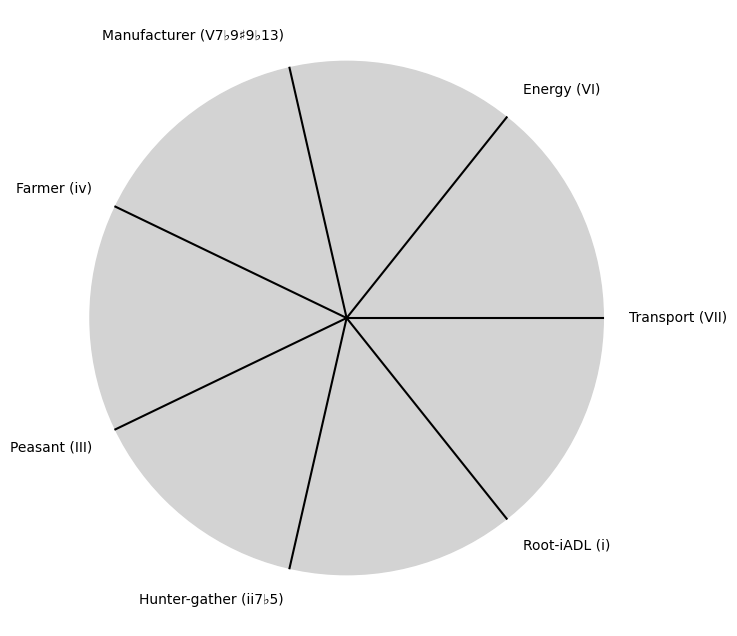
i \(\%\) Overcoming, Self, \(df\)#
\(\alpha, \beta, t\) NexToken:
Parametrizedthe processes that are responsible for human behavior as a moiety of “chains” with some “wiggle-room”
Show code cell source
import matplotlib.pyplot as plt
import numpy as np
# Clock settings; f(t) random disturbances making "paradise lost"
clock_face_radius = 1.0
number_of_ticks = 9
tick_labels = [
"Sun-Genomics", "Chlorophyll-Transcriptomics", "Flora-Proteomics", "Animals-Metabolomics",
"Wood-Epigenomics", "Coal-Lipidomics", "Hydrocarbons-Glycomics", "Renewable-Metagenomics", "Nuclear-Phenomics"
]
# Calculate the angles for each tick (in radians)
angles = np.linspace(0, 2 * np.pi, number_of_ticks, endpoint=False)
# Inverting the order to make it counterclockwise
angles = angles[::-1]
# Create figure and axis
fig, ax = plt.subplots(figsize=(8, 8))
ax.set_xlim(-1.2, 1.2)
ax.set_ylim(-1.2, 1.2)
ax.set_aspect('equal')
# Draw the clock face
clock_face = plt.Circle((0, 0), clock_face_radius, color='lightgrey', fill=True)
ax.add_patch(clock_face)
# Draw the ticks and labels
for angle, label in zip(angles, tick_labels):
x = clock_face_radius * np.cos(angle)
y = clock_face_radius * np.sin(angle)
# Draw the tick
ax.plot([0, x], [0, y], color='black')
# Positioning the labels slightly outside the clock face
label_x = 1.1 * clock_face_radius * np.cos(angle)
label_y = 1.1 * clock_face_radius * np.sin(angle)
# Adjusting label alignment based on its position
ha = 'center'
va = 'center'
if np.cos(angle) > 0:
ha = 'left'
elif np.cos(angle) < 0:
ha = 'right'
if np.sin(angle) > 0:
va = 'bottom'
elif np.sin(angle) < 0:
va = 'top'
ax.text(label_x, label_y, label, horizontalalignment=ha, verticalalignment=va, fontsize=10)
# Remove axes
ax.axis('off')
# Show the plot
plt.show()
Show code cell output
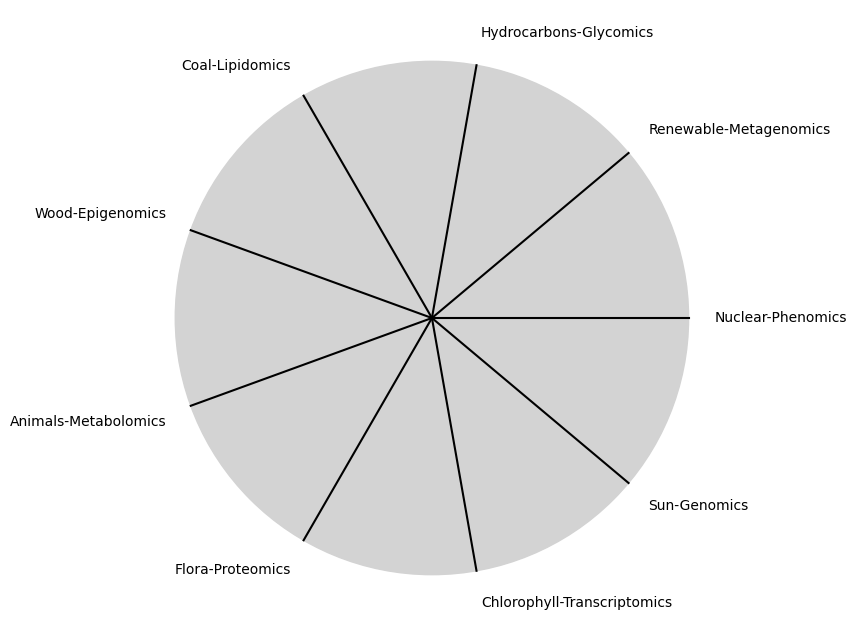
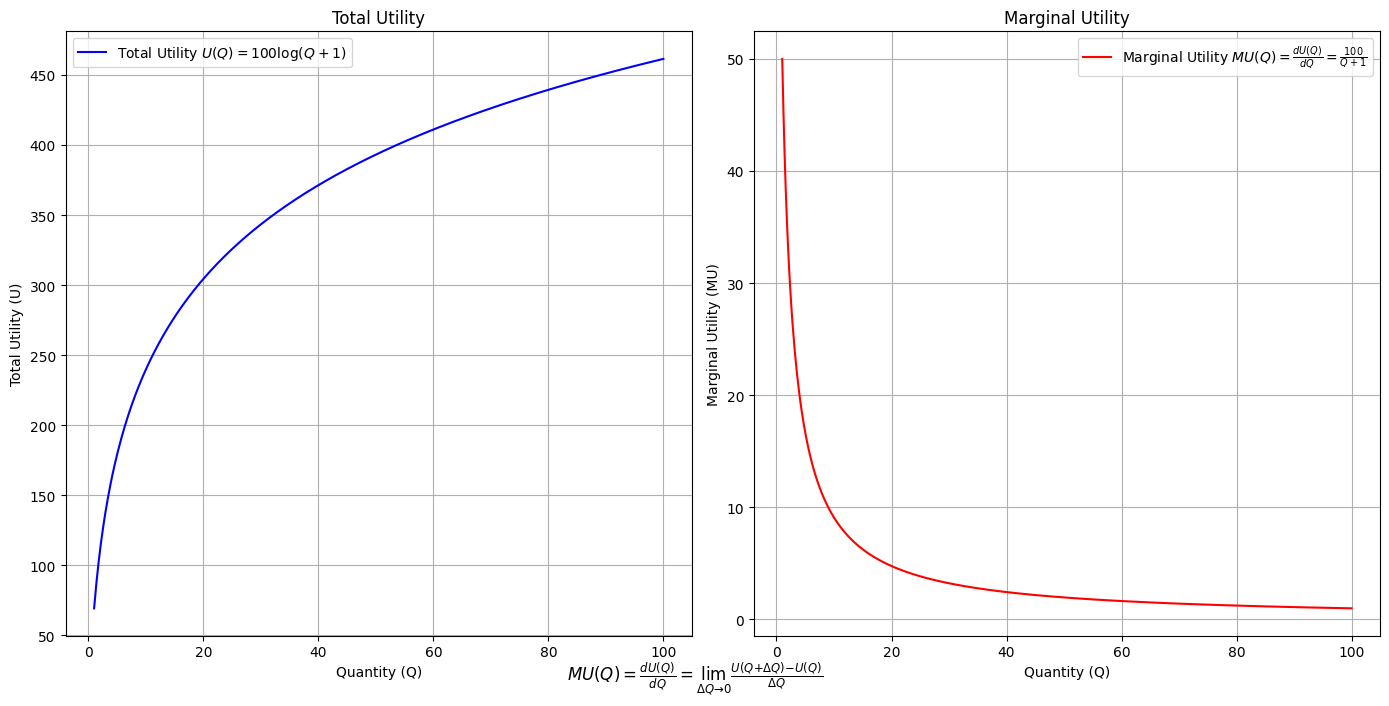
\(SV_t'\) Emotion:
Ultimategoal of life, where all the outlined elements converge. It’s the subjective experience or illusion that life should evoke, the connection between god, neighbor, and self.Thusminor chords may represent “loose” chains whereas dom7 & half-dim are somewhat “tight” chains constraining us to our sense of the nextoken
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
# Define the total utility function U(Q)
def total_utility(Q):
return 100 * np.log(Q + 1) # Logarithmic utility function for illustration
# Define the marginal utility function MU(Q)
def marginal_utility(Q):
return 100 / (Q + 1) # Derivative of the total utility function
# Generate data
Q = np.linspace(1, 100, 500) # Quantity range from 1 to 100
U = total_utility(Q)
MU = marginal_utility(Q)
# Plotting
plt.figure(figsize=(14, 7))
# Plot Total Utility
plt.subplot(1, 2, 1)
plt.plot(Q, U, label=r'Total Utility $U(Q) = 100 \log(Q + 1)$', color='blue')
plt.title('Total Utility')
plt.xlabel('Quantity (Q)')
plt.ylabel('Total Utility (U)')
plt.legend()
plt.grid(True)
# Plot Marginal Utility
plt.subplot(1, 2, 2)
plt.plot(Q, MU, label=r'Marginal Utility $MU(Q) = \frac{dU(Q)}{dQ} = \frac{100}{Q + 1}$', color='red')
plt.title('Marginal Utility')
plt.xlabel('Quantity (Q)')
plt.ylabel('Marginal Utility (MU)')
plt.legend()
plt.grid(True)
# Adding some calculus notation and Greek symbols
plt.figtext(0.5, 0.02, r"$MU(Q) = \frac{dU(Q)}{dQ} = \lim_{\Delta Q \to 0} \frac{U(Q + \Delta Q) - U(Q)}{\Delta Q}$", ha="center", fontsize=12)
plt.tight_layout()
plt.show()
Show code cell output
Show code cell source
import matplotlib.pyplot as plt
import numpy as np
from matplotlib.cm import ScalarMappable
from matplotlib.colors import LinearSegmentedColormap, PowerNorm
def gaussian(x, mean, std_dev, amplitude=1):
return amplitude * np.exp(-0.9 * ((x - mean) / std_dev) ** 2)
def overlay_gaussian_on_line(ax, start, end, std_dev):
x_line = np.linspace(start[0], end[0], 100)
y_line = np.linspace(start[1], end[1], 100)
mean = np.mean(x_line)
y = gaussian(x_line, mean, std_dev, amplitude=std_dev)
ax.plot(x_line + y / np.sqrt(2), y_line + y / np.sqrt(2), color='red', linewidth=2.5)
fig, ax = plt.subplots(figsize=(10, 10))
intervals = np.linspace(0, 100, 11)
custom_means = np.linspace(1, 23, 10)
custom_stds = np.linspace(.5, 10, 10)
# Change to 'viridis' colormap to get gradations like the older plot
cmap = plt.get_cmap('viridis')
norm = plt.Normalize(custom_stds.min(), custom_stds.max())
sm = ScalarMappable(cmap=cmap, norm=norm)
sm.set_array([])
median_points = []
for i in range(10):
xi, xf = intervals[i], intervals[i+1]
x_center, y_center = (xi + xf) / 2 - 20, 100 - (xi + xf) / 2 - 20
x_curve = np.linspace(custom_means[i] - 3 * custom_stds[i], custom_means[i] + 3 * custom_stds[i], 200)
y_curve = gaussian(x_curve, custom_means[i], custom_stds[i], amplitude=15)
x_gauss = x_center + x_curve / np.sqrt(2)
y_gauss = y_center + y_curve / np.sqrt(2) + x_curve / np.sqrt(2)
ax.plot(x_gauss, y_gauss, color=cmap(norm(custom_stds[i])), linewidth=2.5)
median_points.append((x_center + custom_means[i] / np.sqrt(2), y_center + custom_means[i] / np.sqrt(2)))
median_points = np.array(median_points)
ax.plot(median_points[:, 0], median_points[:, 1], '--', color='grey')
start_point = median_points[0, :]
end_point = median_points[-1, :]
overlay_gaussian_on_line(ax, start_point, end_point, 24)
ax.grid(True, linestyle='--', linewidth=0.5, color='grey')
ax.set_xlim(-30, 111)
ax.set_ylim(-20, 87)
# Create a new ScalarMappable with a reversed colormap just for the colorbar
cmap_reversed = plt.get_cmap('viridis').reversed()
sm_reversed = ScalarMappable(cmap=cmap_reversed, norm=norm)
sm_reversed.set_array([])
# Existing code for creating the colorbar
cbar = fig.colorbar(sm_reversed, ax=ax, shrink=1, aspect=90)
# Specify the tick positions you want to set
custom_tick_positions = [0.5, 5, 8, 10] # example positions, you can change these
cbar.set_ticks(custom_tick_positions)
# Specify custom labels for those tick positions
custom_tick_labels = ['5', '3', '1', '0'] # example labels, you can change these
cbar.set_ticklabels(custom_tick_labels)
# Label for the colorbar
cbar.set_label(r'♭', rotation=0, labelpad=15, fontstyle='italic', fontsize=24)
# Label for the colorbar
cbar.set_label(r'♭', rotation=0, labelpad=15, fontstyle='italic', fontsize=24)
cbar.set_label(r'♭', rotation=0, labelpad=15, fontstyle='italic', fontsize=24)
# Add X and Y axis labels with custom font styles
ax.set_xlabel(r'Principal Component', fontstyle='italic')
ax.set_ylabel(r'Emotional State', rotation=0, fontstyle='italic', labelpad=15)
# Add musical modes as X-axis tick labels
# musical_modes = ["Ionian", "Dorian", "Phrygian", "Lydian", "Mixolydian", "Aeolian", "Locrian"]
greek_letters = ['α', 'β','γ', 'δ', 'ε', 'ζ', 'η'] # 'θ' , 'ι', 'κ'
mode_positions = np.linspace(ax.get_xlim()[0], ax.get_xlim()[1], len(greek_letters))
ax.set_xticks(mode_positions)
ax.set_xticklabels(greek_letters, rotation=0)
# Add moods as Y-axis tick labels
moods = ["flow", "control", "relaxed", "bored", "apathy","worry", "anxiety", "arousal"]
mood_positions = np.linspace(ax.get_ylim()[0], ax.get_ylim()[1], len(moods))
ax.set_yticks(mood_positions)
ax.set_yticklabels(moods)
# ... (rest of the code unchanged)
plt.tight_layout()
plt.show()
Show code cell output
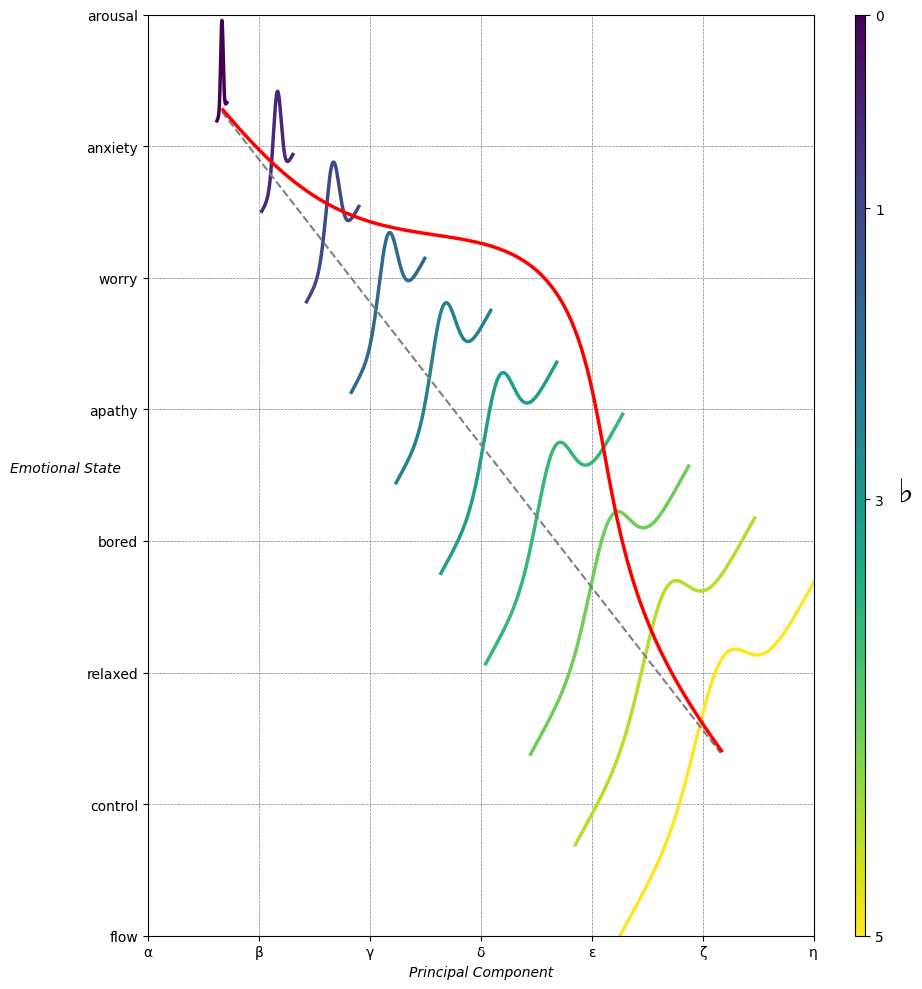

Emotion & Affect as Outcomes & Freewill. And the predictors \(\beta\) are MQ-TEA: Modes (ionian, dorian, phrygian, lydian, mixolydian, locrian), Qualities (major, minor, dominant, suspended, diminished, half-dimished, augmented), Tensions (7th), Extensions (9th, 11th, 13th), and Alterations (♯, ♭) 49#
1. f(t)
\
2. S(t) -> 4. Nxb:t(X'X).X'Y -> 5. b -> 6. df
/
3. h(t)