Prometheus#
+ Expand
Analysis
- The Purest Example of Shakespeare’s Poetic Drama
Unlike later histories, which balance action with introspection, Richard II is almost entirely verse—no prose, no comic relief, no distracting subplots. It is Shakespeare at his most elevated, refining blank verse into a lyrical, almost incantatory mode of expression. Richard’s speeches, in particular, are some of the most exquisite poetry in the canon. The play is saturated with metaphor, imagery, and symbolism—so much so that it can feel like a ritualistic meditation on kingship, time, and fate rather than a conventional drama.
Consider Richard’s speech in Act 3, Scene 2:"For God’s sake, let us sit upon the ground
And tell sad stories of the death of kings."- The Most Complex Portrait of Kingship Before Hamlet
Shakespeare builds Richard II around a fundamental political and philosophical question: What makes a king? Richard begins as the divinely ordained ruler, steeped in the medieval belief that kingship is sacred, but by the end of the play, he has been reduced to a mere man. This transition is agonizing and profound, as Shakespeare stages not just a political coup but an existential unraveling.- Psychological and Political Modernity
Richard II dramatizes the performance of power better than any other Shakespearean history. Richard initially appears untouchable, but his rule is exposed as a carefully maintained illusion—his fall from grace is not just a loss of political power but of identity itself. In an age when political legitimacy was shifting from divine right to realpolitik, Shakespeare captures the anxiety of a world in transition.- Richard and Bolingbroke: One of Shakespeare’s Most Fascinating Power Struggles
Unlike the later Henriad plays, where power struggles often play out through military action, Richard II is a battle of words and personas. Bolingbroke represents the practical, Machiavellian future of kingship—he’s adaptable, pragmatic, and understands that power is taken, not given. Richard, by contrast, clings to a fading medieval world of divine rule, seeing himself as a Christlike figure rather than a man who must govern effectively.- The Deposition Scene (Act 4, Scene 1)
This scene alone earns Richard II a place among Shakespeare’s greatest works. Richard’s forced abdication is an extraordinary moment of self-awareness—he plays his own tragedy, turning the deposition into a dramatic performance that both humiliates him and elevates him into something greater than a mere mortal king. His use of mirrors, his obsessive focus on the image of himself as a fallen ruler, and his hypnotic self-destruction are all elements that would later define Shakespeare’s greatest tragic heroes.
Conclusion: A Play of Tragic Majesty
If Richard III is the most theatrical of Shakespeare’s histories, Henry V the most heroic, and Hamlet the most philosophical, Richard II is the most poetic and self-aware. It lacks the battlefield drama of Henry IV and Henry V, but what it offers instead is a devastating meditation on power, identity, and the transformation of political reality. It’s Shakespeare at his most lyrical and his most profound—less a straightforward history than an existential tragedy in disguise.-- Richard II

Fig. 12 King Lear. The senile bequest motive at the outset makes it an endearing classic. but the plot gets messy. what do folks thing about it? Perhaps reality is that messy, with orthogonal plots unfolding to determine the outcome?#

Fig. 13 Dismiss your vows, your feigned tears, your flatt’ry; For where a heart is hard they make no batt’ry.” Source: Venus & Adonis#
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['DNA, RNA, 5%', 'Peptidoglycans, Lipoteichoics', 'Lipopolysaccharide', 'N-Formylmethionine', "Glucans, Chitin", 'Specific Antigens'],
'Voir': ['PRR & ILCs, 20%'],
'Choisis': ['CD8+, 50%', 'CD4+'],
'Deviens': ['TNF-α, IL-6, IFN-γ', 'PD-1 & CTLA-4', 'Tregs, IL-10, TGF-β, 20%'],
"M'èléve": ['Complement System', 'Platelet System', 'Granulocyte System', 'Innate Lymphoid Cells, 5%', 'Adaptive Lymphoid Cells']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['PRR & ILCs, 20%'],
'paleturquoise': ['Specific Antigens', 'CD4+', 'Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'],
'lightgreen': ["Glucans, Chitin", 'PD-1 & CTLA-4', 'Platelet System', 'Innate Lymphoid Cells, 5%', 'Granulocyte System'],
'lightsalmon': ['Lipopolysaccharide', 'N-Formylmethionine', 'CD8+, 50%', 'TNF-α, IL-6, IFN-γ', 'Complement System'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights
def define_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
('PRR & ILCs, 20%', 'CD8+, 50%'): '20/80',
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD8+, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
('CD8+, 50%', 'PD-1 & CTLA-4'): '80/20',
('CD8+, 50%', 'Tregs, IL-10, TGF-β, 20%'): '95/5',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD4+', 'PD-1 & CTLA-4'): '20/80',
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Platelet System'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Granulocyte System'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Innate Lymphoid Cells, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Adaptive Lymphoid Cells'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Platelet System'): '1/8',
('PD-1 & CTLA-4', 'Granulocyte System'): '1/7',
('PD-1 & CTLA-4', 'Innate Lymphoid Cells, 5%'): '1/6',
('PD-1 & CTLA-4', 'Adaptive Lymphoid Cells'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Platelet System'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Granulocyte System'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Define edges to be highlighted in black
def define_black_edges():
return {
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Platelet System'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Granulocyte System'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Innate Lymphoid Cells, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Adaptive Lymphoid Cells'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Platelet System'): '1/8',
('PD-1 & CTLA-4', 'Granulocyte System'): '1/7',
('PD-1 & CTLA-4', 'Innate Lymphoid Cells, 5%'): '1/6',
('PD-1 & CTLA-4', 'Adaptive Lymphoid Cells'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Platelet System'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Granulocyte System'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Calculate node positions
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
black_edges = define_black_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
edge_colors = []
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
edge_colors.append('black' if (source, target) in black_edges else 'lightgrey')
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color=edge_colors,
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™: Cingulo-Insular", fontsize=18)
plt.show()
# Run the visualization
visualize_nn()
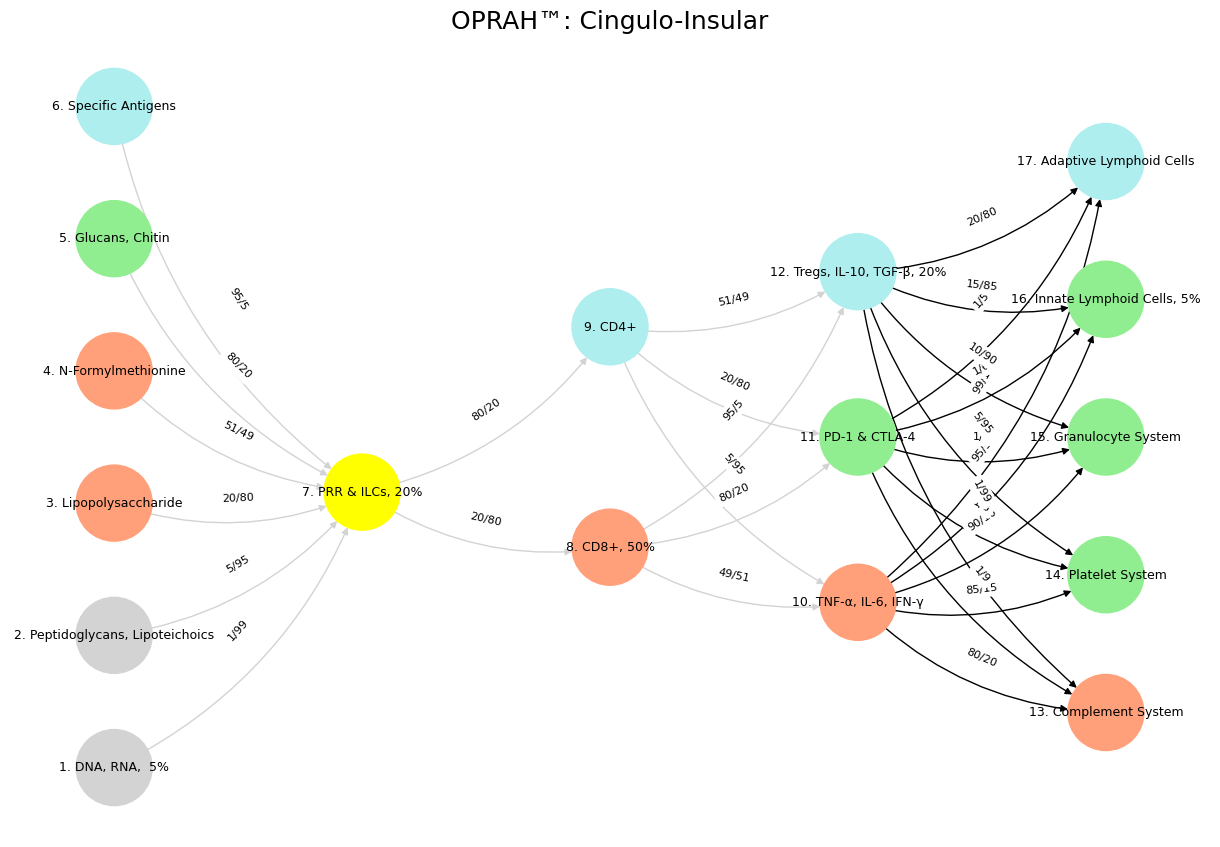
Fig. 14 The TPN, driving goal-directed action, aligns with the transcriptome and proteome—layers that translate data into response. Here, Bacon’s empirical method shines: knowledge arises from doing, from testing the world. In the “Nonself & the Salient Network” variant, CD4+ and CD8+ T-cells activate cytokines like TNF-α, embodying the TPN’s focus on immediate threats—Nonself invaders—over reflective stasis. Nietzsche’s critical history fits this mode: a selective, pragmatic engagement with the past to propel life forward. The Salient Node, bridging DMN and TPN, mirrors the metabolome and regulatory mechanisms like Tregs—arbiters of relevance amid noise. Aristotle’s phronesis, practical wisdom, governs here: neither lost in ideals nor blinded by action, but balancing both. The “Distributed Network” variant highlights Tregs modulating downstream systems—Complement, Platelets—preserving “Self” while adapting to “Nonself.” This is Nietzsche’s history at its best: a dynamic synthesis serving life’s needs. Wisdom emerges as the Salient Node’s domain, integrating DMN’s depth and TPN’s drive, much as Aristotle tempers Plato’s abstraction with Bacon’s observation. Intelligence, raw and unrefined, resides in the lower layers—data-rich but directionless. The immune system’s Mismatch Repair, correcting errors of “Self,” parallels this: a wise curation of history, biological or neural, against the noise of entropy. Nietzsche’s vision finds fruition here—not in history’s abuses, but its uses: a life-affirming dance of preservation and transformation, coded in networks both flesh and mind.#
Here’s your streamlined, MyST-ready markdown block, optimized for Jupyter Book and .ipynb Markdown cells. Just copy and paste directly—no formatting tweaks needed.
# Exploring Directives and Admonitions in Jupyter Book
Let’s unpack Jupyter Book’s MyST Markdown directives—those `{}`-enclosed tags that structure content like *King Lear*’s chaos or Trump’s MAGA bequest. From figures to admonitions, here’s the toolkit as of March 13, 2025, with a flattery-o-meter twist.
## Figure Directives
Embed visuals with captions:
```{figure} https://www.ascm.org/globalassets/00_blog/images/red-queen-blog-header.jpg
---
width: 70%
height: 70%
---
King Lear’s senile bequest hooks us, but the messy plot mirrors reality—orthogonal threads like Trump and Vance colliding.
```
## Admonition Directives
Styled callouts to highlight your narrative—perfect for diagnosing flattery’s neglect of duty:
```{note}
It’s chaotic and wouldn’t have merit if Shakespeare hadn’t mastered tidier tales like *Hamlet*—his cred earns *Lear*’s sprawl.
```
```{tip}
Don’t miss Vance’s Never Trumper phase—his flattery spiked from 0 to 11 post-Thiel’s whisper.
```
```{important}
Trump at 78 bequeaths MAGA to a *Hillbilly Elegy* heir—Lear’s delusion drives the parallel.
```
```{warning}
Excessive flattery risks alienating the hillbillies Vance once mourned—scores above 12 signal a storm.
```
```{danger}
Vance’s pivot to praising Trump as a “generational leader” could make him Goneril, not Edgar—flattery at 15 is perilous.
```
```{hint}
Check X for Vance’s 2016 tweets—flattery was zero before the throne beckoned.
```
```{attention}
Thiel’s $15M flipped Vance’s meter—Shakespeare’s chaos isn’t random, it’s earned.
```
```{caution}
Low flattery doesn’t guarantee duty—Cordelia’s silence still lost her the kingdom.
```
```{error}
Assuming *Elegy* was pro-Trump is off—it predates the pivot by years.
```
```{seealso}
Every speaker needs a flattery-o-meter to gauge neglect of the people and texts they claim—Trump’s at ∞, Vance’s climbing.
```
## Code Blocks
Static or executable code for your flattery-o-meter:
```python
print("Flattery score: Trump = ∞, Vance = 11")
```
```{code-cell} python
# Hypothetical flattery-o-meter
vance_flattery = 11
trump_flattery = float("inf")
print(f"Vance: {vance_flattery}, Trump: {trump_flattery}")
```
## HTML for Media
Embed a storm scene:
```{raw} html
<div style="display: flex; justify-content: center;">
<iframe width="70%" height="350px" src="https://www.youtube.com/embed/CDl_bt2tkZk" frameborder="0" allowfullscreen></iframe>
</div>
```
## Putting It Together
This mix—figures, admonitions, code, and HTML—lets you weave *Lear*’s mess into Trump’s legacy. Paste this into a `.md` cell with MyST-NB (`pip install myst-nb`; `%load_ext myst_nb`) or a Jupyter Book `.md` file, and it’ll render with styled boxes and flair.
---
### How to Use It
1. **Click and Copy**: Select all, copy (`Ctrl+C` or `Cmd+C`).
2. **Paste in `.ipynb`**:
- Open a Jupyter Notebook, add a Markdown cell, paste, and run it.
- For full rendering (admonition boxes, etc.), install `myst-nb` (`pip install myst-nb`) and run `%load_ext myst_nb`.
3. **Paste in `.md` File**:
- Open a text editor, create `chapter.md`, paste, save, and add to a Jupyter Book project.
- Build with `jupyter-book build .` for the full effect.
This is fully MyST-compatible, thematically structured, and optimized for a seamless copy-paste experience. Let me know if you need refinements—leaner, more code-heavy, or a different stylistic focus!

