Life ⚓️#
+ Expand
- What makes for a suitable problem for AI (Demis Hassabis, Nobel Lecture)?
- Space: Massive combinatorial search space
- Function: Clear objective function (metric) to optimize against
- Time: Either lots of data and/or an accurate and efficient simulator
- Guess what else fits the bill (Yours truly, amateur philosopher)?
- Space
- Intestines/villi
- Lungs/bronchioles
- Capillary trees
- Network of lymphatics
- Dendrites in neurons
- Tree branches
- Function
- Energy
- Aerobic respiration
- Delivery to "last mile" (minimize distance)
- Response time (minimize)
- Information
- Exposure to sunlight for photosynthesis
- Time
- Nourishment
- Gaseous exchange
- Oxygen & Nutrients (Carbon dioxide & "Waste")
- Surveillance for antigens
- Coherence of functions
- Water and nutrients from soil
Eco Green’s Strategic Methodology, Innovation, and GESI Approach#
Indicative Methodology: Scientific Excellence in Action#
Eco Green’s approach is anchored in precision ecological engineering—a methodology that integrates soil microbiome rehabilitation, adaptive agronomic modeling, and farmer-centric implementation strategies. The project will employ multi-tiered field trials to benchmark the efficacy of its patented organic liquid fertilizer against synthetic inputs, focusing on soil regeneration, crop yield enhancement, and economic feasibility.

Fig. 4 Veni-Vidi, Veni-Vidi-Vici. Yep, Red Queen Hypothesis all the way. This essay integrates your model’s adversarial framework, immune system parallels, and combinatorial search logic to explore racism as an error-prone interrogation mechanism. Source: Negotiated Identity#
Key activities aligned with project outputs:
Soil Regeneration & Microbial Analysis: Longitudinal studies will evaluate the impact of Eco Green fertilizer on soil carbon sequestration, microbial biodiversity, and water retention.
Crop-Specific Optimization Trials: Research will establish ideal application rates across staple crops like teff, maize, and coffee.
Economic Viability Studies: Comparative ROI analyses will assess the financial advantage of organic inputs over conventional fertilizers for smallholder farmers.
Adoption Acceleration Program: On-farm demonstrations, digital advisory tools, and government extension partnerships will drive behavioral change toward sustainable agricultural practices.
Climate Resilience and Policy Integration: The project will quantify the fertilizer’s GHG reduction potential, strengthening Ethiopia’s climate mitigation policies and bolstering local fertilizer sovereignty.
Utilizing state-of-the-art fermentation biotechnology, Eco Green’s organic liquid fertilizers will be systematically tested in controlled trials and real-world farming conditions, leveraging partnerships with universities, research institutions, and agricultural cooperatives. The results will be disseminated through scientific publications, policy briefs, and farmer training materials, ensuring a broad adoption curve and high-impact scalability.
Innovation: Disrupting the Chemical Dependency Cycle#
Eco Green’s innovative edge lies in its ability to biologically reprogram agricultural resilience. This is achieved through:
A patented organic fermentation process that enriches soil health while outcompeting chemical fertilizers in cost-effectiveness.
Microbial Activation Technology (MAT): A bio-enhanced formulation that restores microbial diversity, triggering a self-reinforcing soil fertility cycle.
Farmer-Led Fertilizer Sovereignty Model: Moving beyond subsidy dependency, Eco Green introduces a cooperative production and distribution system that positions farmers as primary stakeholders in organic input manufacturing.
Policy Interventions & Digital Farmer Training: Eco Green integrates a QR-coded knowledge hub where smallholder farmers access customized guidance on soil nutrition, organic inputs, and market linkages.
By challenging Ethiopia’s reliance on imported synthetic fertilizers, Eco Green reprograms the agricultural supply chain, reducing foreign currency outflows and empowering local production.
Conclusion: Immunizing Ethiopian Agriculture Against Synthetic Dependency#
Eco Green is not just a fertilizer company—it is an immune response mechanism for Ethiopia’s biological, economic, and climate resilience. By embedding scientific rigor, technological innovation, and equity-driven implementation, the project redefines fertilizer sovereignty, ensuring Ethiopia’s soil remembers itself—not as a dependent system, but as a self-sustaining, regenerative force.
Show code cell source
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
# Define the neural network layers
def define_layers():
return {
'Suis': ['DNA, RNA, 5%', 'Peptidoglycans, Lipoteichoics', 'Lipopolysaccharide', 'N-Formylmethionine', "Glucans, Chitin", 'Specific Antigens'],
'Voir': ['PRR & ILCs, 20%'],
'Choisis': ['CD8+, 50%', 'CD4+'],
'Deviens': ['TNF-α, IL-6, IFN-γ', 'PD-1 & CTLA-4', 'Tregs, IL-10, TGF-β, 20%'],
"M'èléve": ['Complement System', 'Platelet System', 'Granulocyte System', 'Innate Lymphoid Cells, 5%', 'Adaptive Lymphoid Cells']
}
# Assign colors to nodes
def assign_colors():
color_map = {
'yellow': ['PRR & ILCs, 20%'],
'paleturquoise': ['Specific Antigens', 'CD4+', 'Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'],
'lightgreen': ["Glucans, Chitin", 'PD-1 & CTLA-4', 'Platelet System', 'Innate Lymphoid Cells, 5%', 'Granulocyte System'],
'lightsalmon': ['Lipopolysaccharide', 'N-Formylmethionine', 'CD8+, 50%', 'TNF-α, IL-6, IFN-γ', 'Complement System'],
}
return {node: color for color, nodes in color_map.items() for node in nodes}
# Define edge weights
def define_edges():
return {
('DNA, RNA, 5%', 'PRR & ILCs, 20%'): '1/99',
('Peptidoglycans, Lipoteichoics', 'PRR & ILCs, 20%'): '5/95',
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
('PRR & ILCs, 20%', 'CD8+, 50%'): '20/80',
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD8+, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
('CD8+, 50%', 'PD-1 & CTLA-4'): '80/20',
('CD8+, 50%', 'Tregs, IL-10, TGF-β, 20%'): '95/5',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD4+', 'PD-1 & CTLA-4'): '20/80',
('CD4+', 'Tregs, IL-10, TGF-β, 20%'): '51/49',
('TNF-α, IL-6, IFN-γ', 'Complement System'): '80/20',
('TNF-α, IL-6, IFN-γ', 'Platelet System'): '85/15',
('TNF-α, IL-6, IFN-γ', 'Granulocyte System'): '90/10',
('TNF-α, IL-6, IFN-γ', 'Innate Lymphoid Cells, 5%'): '95/5',
('TNF-α, IL-6, IFN-γ', 'Adaptive Lymphoid Cells'): '99/1',
('PD-1 & CTLA-4', 'Complement System'): '1/9',
('PD-1 & CTLA-4', 'Platelet System'): '1/8',
('PD-1 & CTLA-4', 'Granulocyte System'): '1/7',
('PD-1 & CTLA-4', 'Innate Lymphoid Cells, 5%'): '1/6',
('PD-1 & CTLA-4', 'Adaptive Lymphoid Cells'): '1/5',
('Tregs, IL-10, TGF-β, 20%', 'Complement System'): '1/99',
('Tregs, IL-10, TGF-β, 20%', 'Platelet System'): '5/95',
('Tregs, IL-10, TGF-β, 20%', 'Granulocyte System'): '10/90',
('Tregs, IL-10, TGF-β, 20%', 'Innate Lymphoid Cells, 5%'): '15/85',
('Tregs, IL-10, TGF-β, 20%', 'Adaptive Lymphoid Cells'): '20/80'
}
# Define edges to be highlighted in black
def define_black_edges():
return {
('Lipopolysaccharide', 'PRR & ILCs, 20%'): '20/80',
('N-Formylmethionine', 'PRR & ILCs, 20%'): '51/49',
("Glucans, Chitin", 'PRR & ILCs, 20%'): '80/20',
('Specific Antigens', 'PRR & ILCs, 20%'): '95/5',
('PRR & ILCs, 20%', 'CD8+, 50%'): '20/80',
('PRR & ILCs, 20%', 'CD4+'): '80/20',
('CD4+', 'TNF-α, IL-6, IFN-γ'): '5/95',
('CD8+, 50%', 'TNF-α, IL-6, IFN-γ'): '49/51',
}
# Calculate node positions
def calculate_positions(layer, x_offset):
y_positions = np.linspace(-len(layer) / 2, len(layer) / 2, len(layer))
return [(x_offset, y) for y in y_positions]
# Create and visualize the neural network graph
def visualize_nn():
layers = define_layers()
colors = assign_colors()
edges = define_edges()
black_edges = define_black_edges()
G = nx.DiGraph()
pos = {}
node_colors = []
# Create mapping from original node names to numbered labels
mapping = {}
counter = 1
for layer in layers.values():
for node in layer:
mapping[node] = f"{counter}. {node}"
counter += 1
# Add nodes with new numbered labels and assign positions
for i, (layer_name, nodes) in enumerate(layers.items()):
positions = calculate_positions(nodes, x_offset=i * 2)
for node, position in zip(nodes, positions):
new_node = mapping[node]
G.add_node(new_node, layer=layer_name)
pos[new_node] = position
node_colors.append(colors.get(node, 'lightgray'))
# Add edges with updated node labels
edge_colors = []
for (source, target), weight in edges.items():
if source in mapping and target in mapping:
new_source = mapping[source]
new_target = mapping[target]
G.add_edge(new_source, new_target, weight=weight)
edge_colors.append('black' if (source, target) in black_edges else 'lightgrey')
# Draw the graph
plt.figure(figsize=(12, 8))
edges_labels = {(u, v): d["weight"] for u, v, d in G.edges(data=True)}
nx.draw(
G, pos, with_labels=True, node_color=node_colors, edge_color=edge_colors,
node_size=3000, font_size=9, connectionstyle="arc3,rad=0.2"
)
nx.draw_networkx_edge_labels(G, pos, edge_labels=edges_labels, font_size=8)
plt.title("OPRAH™: Dorsal", fontsize=18)
plt.show()
# Run the visualization
visualize_nn()
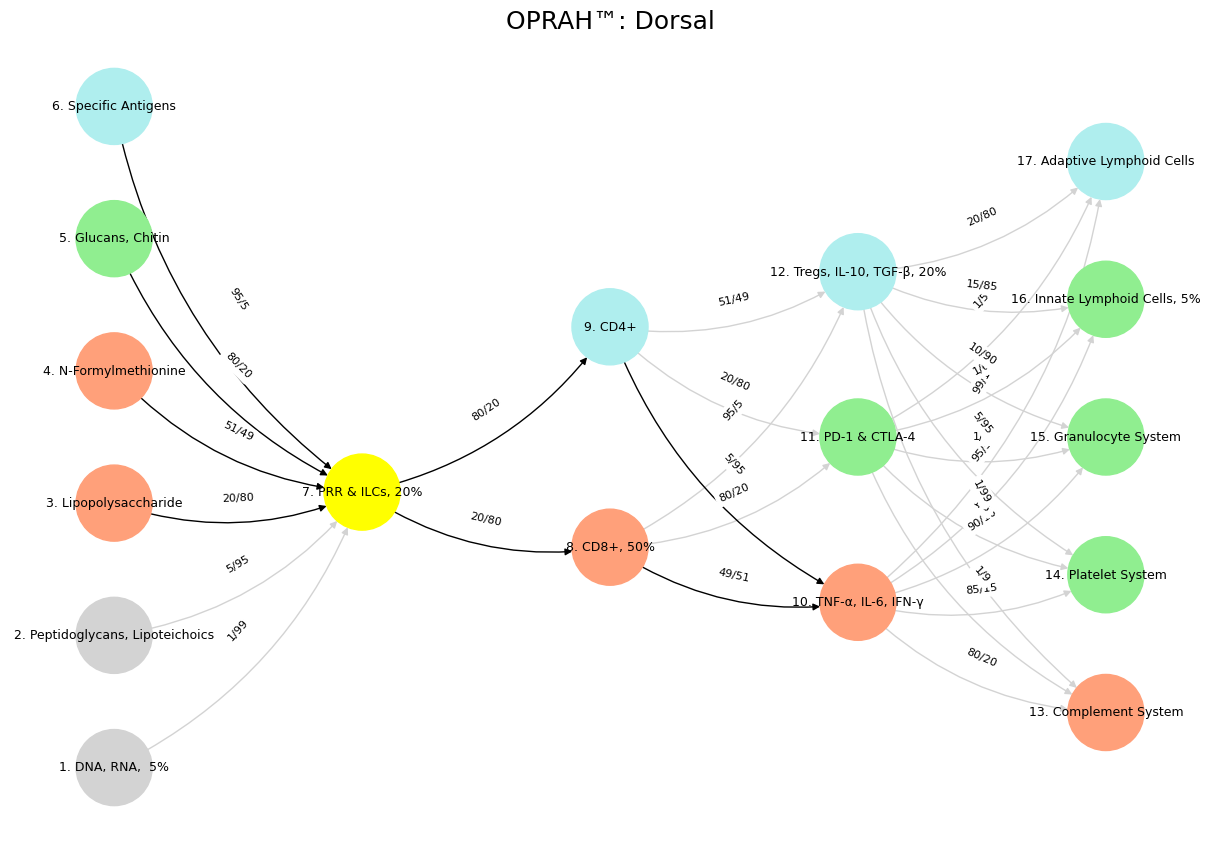

Fig. 5 Innovation: Veni-Vidi, Veni-Vidi-Vici. If you’re protesting then you’re not running fast enough. Thus spake the Red Queens#

