Content with notebooks#
You can also create content with Jupyter Notebooks. This means that you can include code blocks and their outputs in your book.
Markdown + notebooks#
As it is markdown, you can embed images, HTML, etc into your posts!
You can also \(add_{math}\) and
or
But make sure you $Escape $your $dollar signs $you want to keep!
MyST markdown#
MyST markdown works in Jupyter Notebooks as well. For more information about MyST markdown, check out the MyST guide in Jupyter Book, or see the MyST markdown documentation.
Code blocks and outputs#
Jupyter Book will also embed your code blocks and output in your book. For example, here’s some sample Matplotlib code:
from matplotlib import rcParams, cycler
import matplotlib.pyplot as plt
import numpy as np
from scipy import stats
import pandas as pd
plt.ion()
<contextlib.ExitStack at 0x103c6b690>
# Fixing random state for reproducibility
np.random.seed(19680801)
N = 10
data = [np.logspace(0, 1, 100) + np.random.randn(100) + ii for ii in range(N)]
data = np.array(data).T
cmap = plt.cm.coolwarm
rcParams['axes.prop_cycle'] = cycler(color=cmap(np.linspace(0, 1, N)))
from matplotlib.lines import Line2D
custom_lines = [Line2D([0], [0], color=cmap(0.), lw=4),
Line2D([0], [0], color=cmap(.5), lw=4),
Line2D([0], [0], color=cmap(1.), lw=4)]
fig, ax = plt.subplots(figsize=(10, 5))
lines = ax.plot(data)
ax.legend(custom_lines, ['Cold', 'Medium', 'Hot']);
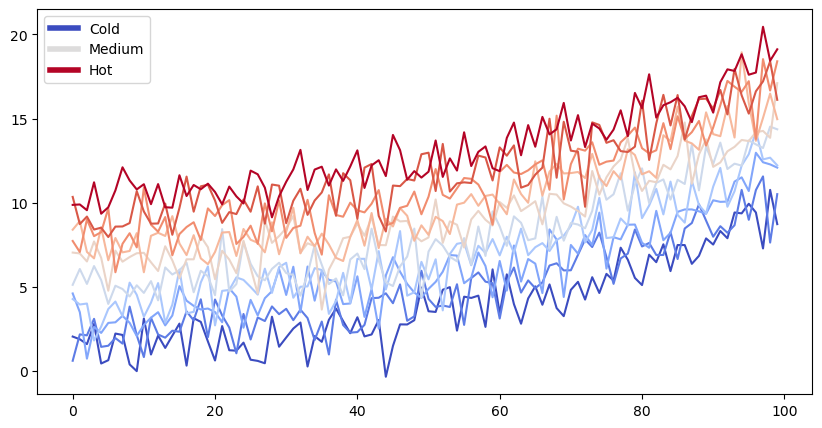
There is a lot more that you can do with outputs (such as including interactive outputs) with your book. For more information about this, see the Jupyter Book documentation
import numpy as np
import matplotlib.pyplot as plt
from scipy import stats
import pandas as pd
# Load data without specifying data types initially
df = pd.read_csv('~/documents/github/csv/donor_live.csv', low_memory=False)
# Print DataFrame info to verify load
print(df.info())
# Clean and convert data types
df['pers_id'] = pd.to_numeric(df['pers_id'], errors='coerce')
df['don_age_in_months'] = pd.to_numeric(df['don_age_in_months'], errors='coerce')
df['don_age'] = pd.to_numeric(df['don_age'], errors='coerce')
df['don_race'] = pd.to_numeric(df['don_race'], errors='coerce')
# Display basic statistics for continuous variables
print(df[['don_age_in_months', 'don_age', 'don_race']].describe())
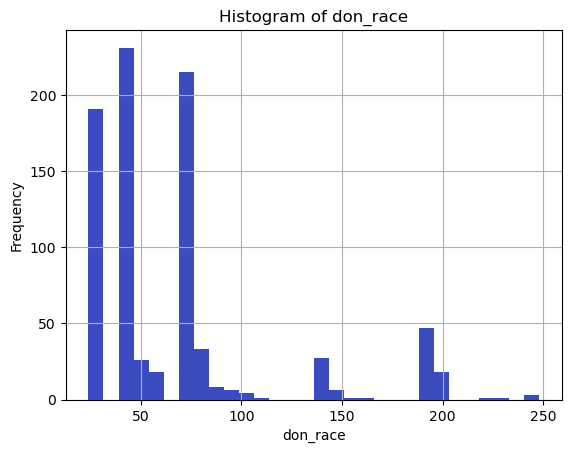
# Plot histograms for continuous variables
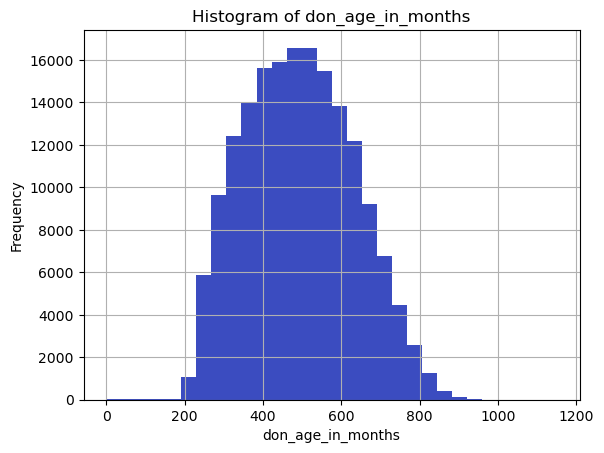
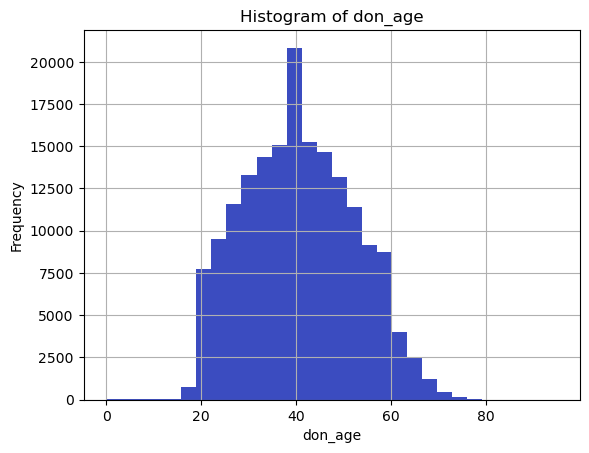
continuous_vars = ['don_age_in_months', 'don_age', 'don_race']
for var in continuous_vars:
plt.figure()
df[var].dropna().hist(bins=30)
plt.title(f'Histogram of {var}')
plt.xlabel(var)
plt.ylabel('Frequency')
plt.show()
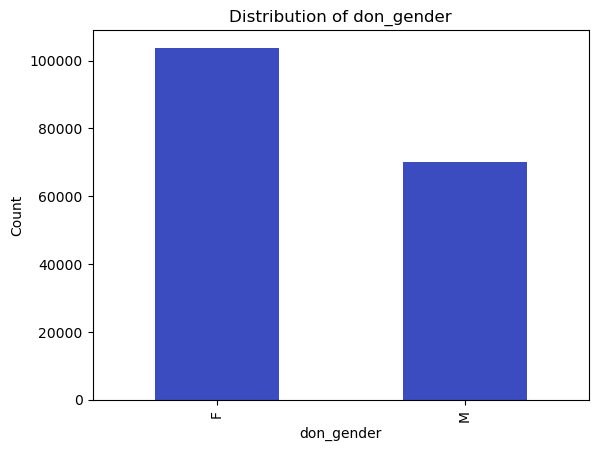
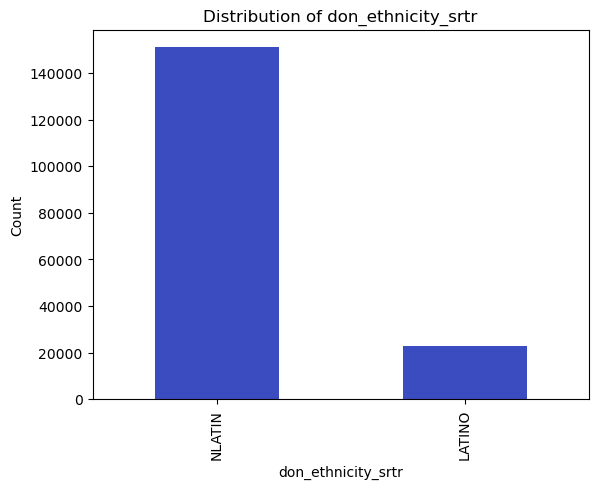
# Count and plot binary/categorical variables
binary_vars = ['don_gender', 'don_ethnicity_srtr']
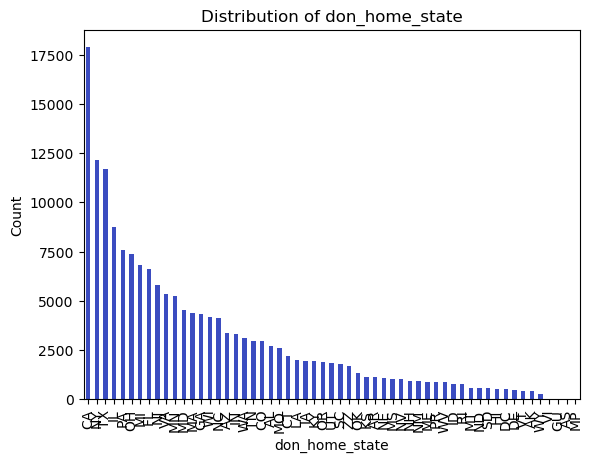
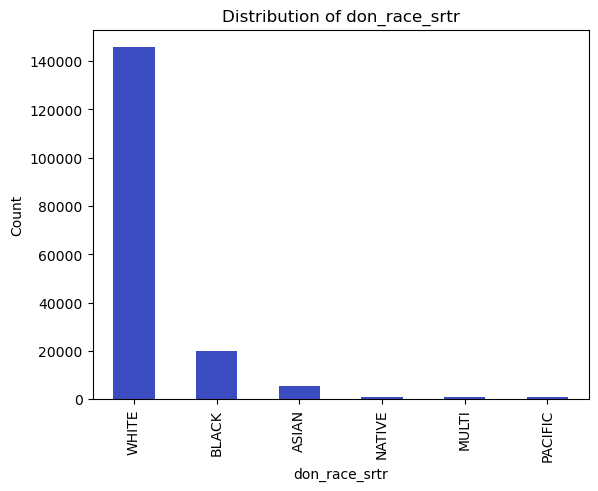
categorical_vars = ['don_ty', 'don_home_state', 'don_race_srtr']
for var in binary_vars + categorical_vars:
plt.figure()
df[var].value_counts().plot(kind='bar')
plt.title(f'Distribution of {var}')
plt.xlabel(var)
plt.ylabel('Count')
plt.show()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 173929 entries, 0 to 173928
Data columns (total 36 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 don_id 173929 non-null object
1 pers_id 173929 non-null int64
2 don_ty 173929 non-null object
3 don_age_in_months 173922 non-null float64
4 don_age 173922 non-null float64
5 don_gender 173929 non-null object
6 don_home_state 170492 non-null object
7 don_ethnicity_pre2004 131260 non-null float64
8 don_race_pre2004 131260 non-null float64
9 don_race 173851 non-null object
10 don_race_srtr 173815 non-null object
11 don_ethnicity_srtr 173929 non-null object
12 don_home_country 0 non-null float64
13 don_hgt_cm 130924 non-null float64
14 don_wgt_kg 132825 non-null float64
15 don_recov_dt 173929 non-null object
16 don_relationship_ty 172919 non-null object
17 don_diab 101127 non-null object
18 don_hyperten_diet 3283 non-null object
19 don_hyperten_diuretics 3283 non-null object
20 don_hyperten_other_meds 3283 non-null object
21 don_ki_creat_preop 128276 non-null float64
22 don_bp_preop_syst 118935 non-null float64
23 don_bp_preop_diast 118915 non-null float64
24 pers_esrd_death_dt 79 non-null float64
25 pers_ssa_death_dt 1395 non-null object
26 don_hyperten_postop 101066 non-null object
27 pers_esrd_first_service_dt 706 non-null float64
28 pers_esrd_first_dial_dt 690 non-null float64
29 pers_esrd_ki_dgn 332 non-null float64
30 don_race_american_indian 173929 non-null int64
31 don_race_asian 173929 non-null int64
32 don_race_black_african_american 173929 non-null int64
33 don_race_hispanic_latino 173929 non-null int64
34 don_race_native_hawaiian 173929 non-null int64
35 don_race_white 173929 non-null int64
dtypes: float64(14), int64(7), object(15)
memory usage: 47.8+ MB
None
don_age_in_months don_age don_race
count 173922.000000 173922.000000 838.000000
mean 493.620244 40.676142 65.250597
std 139.174427 11.600951 48.149922
min 0.000000 0.000000 24.000000
25% 385.000000 32.000000 40.000000
50% 489.000000 40.000000 40.000000
75% 595.000000 49.000000 72.000000
max 1151.000000 95.000000 248.000000
# Read the CSV file into a DataFrame
df = pd.read_csv('~/documents/github/csv/donor_live.csv')
# Show the DataFrame
You can find the dimensions of a DataFrame using the .shape attribute. This attribute returns a tuple representing the dimensions of the DataFrame, where the first element is the number of rows and the second is the number of columns.
Here’s how to find the dimensions of your DataFrame df:
# Get the dimensions of the DataFrame
rows, cols = df.shape
# Print the dimensions
print(f"The DataFrame has {rows} rows and {cols} columns.")
The DataFrame has 173929 rows and 36 columns.
This will give you a quick idea of how large your dataset is.
Next, you can use the .head() method to print the first 5 rows of your DataFrame. This is a good way to get a feel for what information is in your dataset. You can pass a number into the .head() method to print a different number of rows. For example, df.head(10) will print the first 10 rows of your DataFrame.
df.head()
| don_id | pers_id | don_ty | don_age_in_months | don_age | don_gender | don_home_state | don_ethnicity_pre2004 | don_race_pre2004 | don_race | ... | don_hyperten_postop | pers_esrd_first_service_dt | pers_esrd_first_dial_dt | pers_esrd_ki_dgn | don_race_american_indian | don_race_asian | don_race_black_african_american | don_race_hispanic_latino | don_race_native_hawaiian | don_race_white | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | HRG962 | 289 | L | 490.0 | 40.0 | F | TX | 2.0 | 8.0 | 8: White | ... | N | NaN | NaN | NaN | 0 | 0 | 0 | 0 | 0 | 1 |
| 1 | IT5273 | 484 | L | 312.0 | 26.0 | F | OH | 2.0 | 8.0 | 8: White | ... | N | NaN | NaN | NaN | 0 | 0 | 0 | 0 | 0 | 1 |
| 2 | HRL860 | 840 | L | 648.0 | 54.0 | M | ND | 2.0 | 8.0 | 8: White | ... | N | NaN | NaN | NaN | 0 | 0 | 0 | 0 | 0 | 1 |
| 3 | ZWB847 | 967 | L | 564.0 | 47.0 | M | NY | 1.0 | 8.0 | 2000: Hispanic/Latino | ... | NaN | NaN | NaN | NaN | 0 | 0 | 0 | 1 | 0 | 0 |
| 4 | ALB852 | 1145 | L | 326.0 | 27.0 | F | CA | 1.0 | 8.0 | 2000: Hispanic/Latino | ... | NaN | NaN | NaN | NaN | 0 | 0 | 0 | 1 | 0 | 0 |
5 rows × 36 columns
You can restrict the DataFrame to a subset of rows and columns using slicing. Here’s how to keep only the first 1000 rows and the first 9 columns:
# Subset the DataFrame
subset_df = df.iloc[:1000, :9]
# Show the shape of the new DataFrame to confirm
print(f"The shape of the subset DataFrame is {subset_df.shape}")
The shape of the subset DataFrame is (1000, 9)
Explanation:
df.iloc[:1000, :9]uses theilocindexer for Pandas DataFrames. The:1000selects the first 1000 rows and the:9selects the first 9 columns.subset_df.shapewill display the shape of the new DataFrame, which should be (1000, 9) to confirm that you’ve correctly subsetted it.
Now subset_df contains only the first 1000 rows and the first 9 columns of the original df.
subset_df.head()
| don_id | pers_id | don_ty | don_age_in_months | don_age | don_gender | don_home_state | don_ethnicity_pre2004 | don_race_pre2004 | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | HRG962 | 289 | L | 490.0 | 40.0 | F | TX | 2.0 | 8.0 |
| 1 | IT5273 | 484 | L | 312.0 | 26.0 | F | OH | 2.0 | 8.0 |
| 2 | HRL860 | 840 | L | 648.0 | 54.0 | M | ND | 2.0 | 8.0 |
| 3 | ZWB847 | 967 | L | 564.0 | 47.0 | M | NY | 1.0 | 8.0 |
| 4 | ALB852 | 1145 | L | 326.0 | 27.0 | F | CA | 1.0 | 8.0 |
# Select the first 100 rows and cherry-pick columns by index (e.g., columns at index 0, 2, 4, and 6)
subset_csv = df.iloc[:100, [3, 4, 6]]
subset_csv.to_csv('~/documents/github/work/subset.csv', index=False)
# Summary statistics to identify continuous variables
cont_summary = subset_df.describe()
print("Continuous Variables:")
print(cont_summary.columns.tolist()) # These are likely to be continuous variables
# Identify binary variables
binary_vars = [col for col in subset_df.columns if subset_df[col].nunique() == 2]
print("\nBinary Variables:")
print(binary_vars)
# Identify categorical variables
cat_vars = [col for col in subset_df.select_dtypes(include=['object']).columns if col not in binary_vars]
print("\nCategorical Variables:")
print(cat_vars)
Continuous Variables:
['pers_id', 'don_age_in_months', 'don_age', 'don_ethnicity_pre2004', 'don_race_pre2004']
Binary Variables:
['don_gender', 'don_ethnicity_pre2004']
Categorical Variables:
['don_id', 'don_ty', 'don_home_state']
import numpy as np
import matplotlib.pyplot as plt
from scipy import stats
# Sample data (replace with your own)
ages = subset_df['don_age'].dropna() # Remove NaN values if any
# Add jitter to x-values for visualization
np.random.seed(42)
jitter = np.random.normal(0, 0.1, len(ages))
# Scatter plot
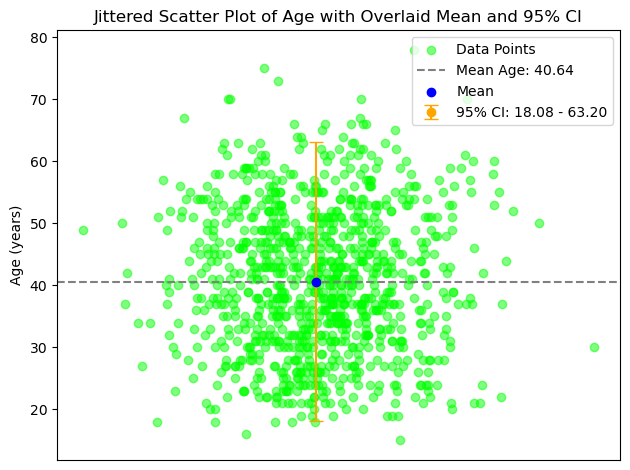
plt.scatter(jitter, ages, color='lime', alpha=0.5, label='Data Points')
# Calculate mean and 95% CI for ages
mean_age = np.mean(ages)
confidence = 0.95
std_dev = np.std(ages)
ci = std_dev * stats.t.ppf((1 + confidence) / 2, len(ages) - 1)
# Overlay mean as a horizontal line (in gray)
plt.axhline(y=mean_age, color='gray', linestyle='--', label=f'Mean Age: {mean_age:.2f}')
# Overlay mean as a blue dot
plt.scatter(0, mean_age, color='blue', zorder=5, label='Mean')
# Overlay 95% CI as a vertical error bar (in orange)
plt.errorbar(0, mean_age, yerr=ci, color='orange', fmt='o', capsize=5, label=f'95% CI: {mean_age - ci:.2f} - {mean_age + ci:.2f}')
# Remove x-axis ticks, labels, and title
plt.xticks([])
plt.xlabel('')
plt.ylabel('Age (years)')
plt.title('Jittered Scatter Plot of Age with Overlaid Mean and 95% CI')
plt.legend()
plt.tight_layout()
plt.show()